- Home
- Member Resources
- Articles
- The Updates of Molecular Testing in T-cell Malignancies
After reading the article, hear more background from the author on the CAPcast, Updates of Molecular Testing in T-Cell Malignancies.
Evolving molecular technologies and genomic knowledge has significantly changed the diagnosis and management of T-cell malignancies. More and more clinical laboratories have adopted next generation sequencing platforms to assess clonality and to identify diagnostic and predictive biomarkers. Understanding these practice changes will facilitate the pathologist’s approach to T-cell malignancies and affect the management of cancer patients.
T-cell Receptor Gene Clonal Rearrangement Studies
T-cell receptor (TCR) clonality assessment is often essential in the initial diagnosis of T-cell lymphomas (T-NHL), especially cutaneous T-cell lymphoma (CTCL), which may be challenging to diagnose. The current gold standard of clonality assessment by fragment analysis has distinct limitations such as low sensitivity and specificity, inability to separate multiple rearrangements with same size, and no or limited utility in the detection of minimal residual disease(MRD) in post treatment samples. Next generation sequencing (NGS) offers unique advantages of differentiating clonal populations sharing the same fragment size and providing specific clonal sequences, which can be tracked in subsequent samples. This allows more accurate initial diagnosis, as well as, highly sensitive detection of MRD in the post treatment settings.
NGS-based assay for characterization of the T cell clonal rearrangement uses primers sets targeting various regions of the T-cell receptor beta or gamma (TRB or TRG) gene.1 The primers contain barcoded sequence adaptors, allowing demultiplexing of reads after sequencing. Sequences are then analyzed though a bioinformatics pipeline. Criteria for clonality calling has been described by groups for IGH and TCR, respectively.2-4 However, there are no standard rules yet. One article suggested that to meet the criterion of being clonal, the top one or two clusters must be more than 4.5% of the total reads and 4-fold high than the background.4 Another published validation study indicated that clonal calls require the top 1 or 2 merged reads to be >2.5% and >5 times of the frequency of the 4th most prevalent clonotype.2 Our unpublished data determined that the criteria to call a clonal sequence should be more strict to avoid false positives, especially in low tumor content or post treatment samples. We use a combination of 5% and 10-fold of the background for clonotype (the manuscript is under preparation). In addition, laboratories that perform both characterization and disease monitoring using NGS assays should avoid cross-contamination during sequencing by setting up the samples from same patient in different runs, using different barcode indexes, on different sequencing instruments whenever possible.

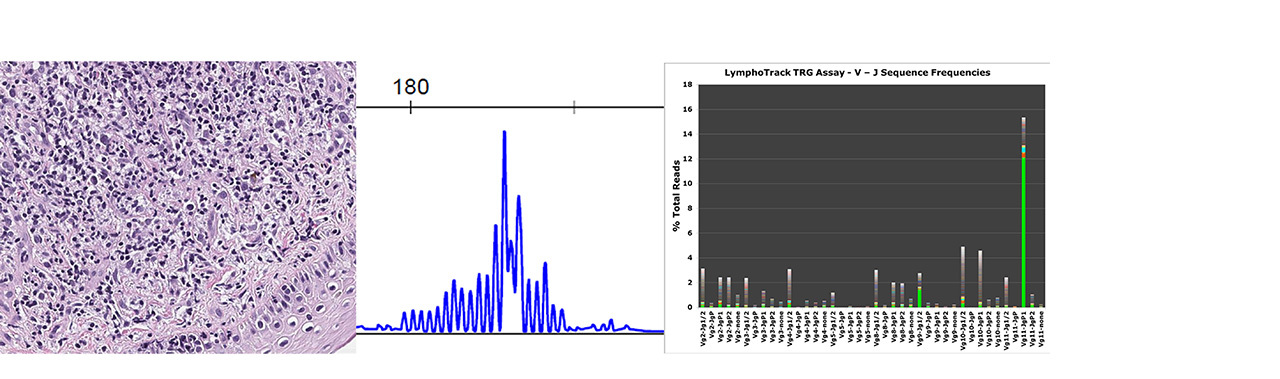
Figure 1. Skin biopsy with Atypical Lymphocytic Infiltrate
A: Morphologic and immunophenotypic features are not diagnostic
B: Fragment analysis showed no clonal rearrangement but a prominent peak
C: NGS detected a clonal sequence with Vg11/JgP1 rearrangement
NGS = Next Generation Sequencing
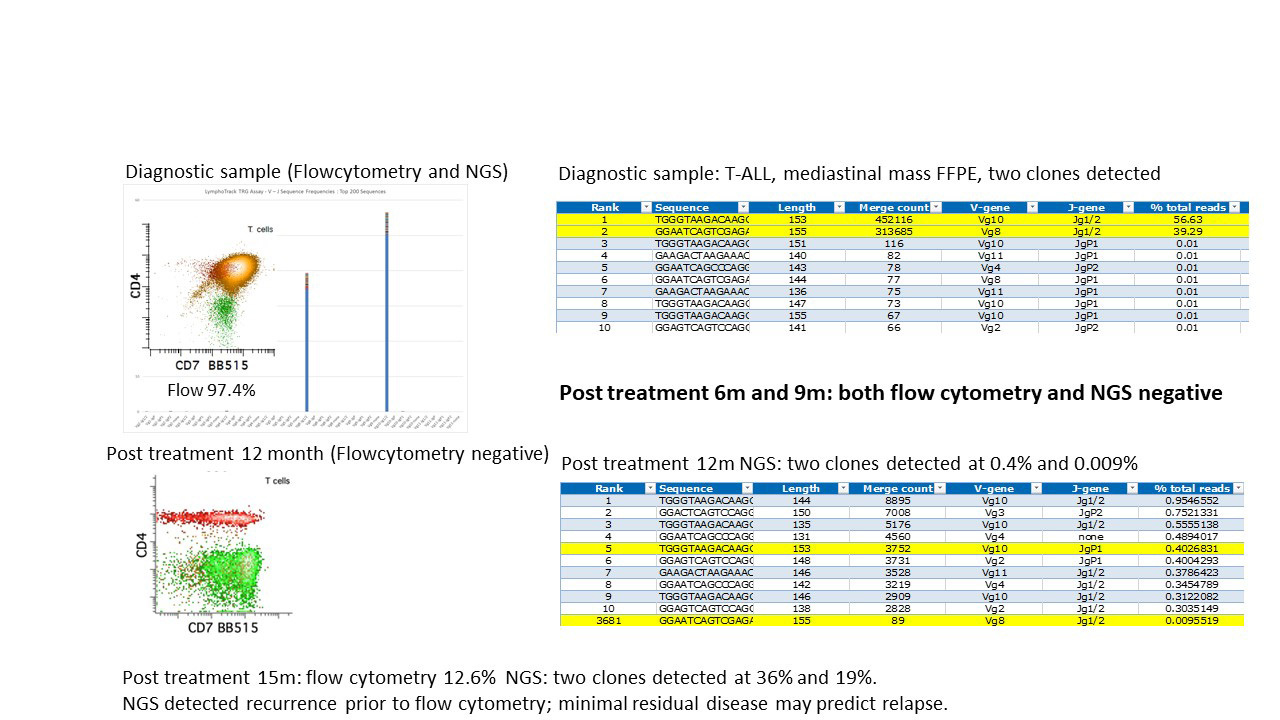
If the same instrument must be used, a rotating instrument schedule should be instituted so that samples from the same patient would not be sequenced within 3 runs of each other. Monitoring studies using NGS has a sensitivity as low as 0.001% (our unpublished data), which may have some value in early detection of relapse.

Figure 2. NGS showed Higher Sensitivity of in Detecting Relapse than Flow Cytometry
NGS = Next Generation Sequencing
The superior sensitivity of clonal rearrangement assessment by NGS over fragment analysis is illustrated in this CTCL (Figure 1), which is a skin biopsy with atypical lymphocytic infiltrate; while the higher sensitivity of NGS detects disease relapse earlier than flow cytometry is demonstrated in Figure 2.
Genomic Profiling of T-cell Leukemia and Lymphomas
With the development and wide applications of next generation sequencing in tumor genomic profiling, the genetic alterations of several subtypes of T-cell leukemias and lymphomas have been discovered in the past a few years.
Both DNA and RNA sequencing are needed to be able to capture the variety of somatic alterations and gene fusions that occur in T-cell malignancies. Most clinical laboratories use a targeted large panel with either hybrid or amplicon capture for DNA sequencing,5,6 and amplicon based, Anchored multiplex-PCR for RNAseq, which can detect unknown fusion and is very promising for the identification of cryptic structural variants.7
T-Lymphoblastic Leukemia/Lymphoma and Early T-cell Precursor Acute Lymphoblastic Leukemia
Abnormal karyotype and activating mutations have been observed in 50-70% of T-lymphoblastic leukemia/lymphoma (T-ALL/LBL). Gene fusions of KMT2A, LMO2, TAL1, TLX1, TLX3, TRA, TRB, TRG, and PICALM-MLL10 are often diagnostic. Activating mutations of FBXW7, NOTCH1, PHF6, and PTEN genes may also assist the diagnosis of T-ALL.8
Early T-cell precursor acute lymphoblastic leukemia (ETP ALL) is an aggressive malignancy of both children and adults. Whole genome sequencing identified somatic mutations of multiple genes including BRAF, DNMT3A, ECT2L, EED, EP300, ETV6, EZH2, FLT3, GATA2, GATA3, IL7R, JAK1, JAK3, KRAS, NRAS, PHF6, RELN, RUNX1, SH2B3, and SUZ12 in ETP ALL.9 Some of these, such as GATA3, DNM2, ECT2L, EZH2, PHF6 and RUNX1, are recurrent mutations and may have some diagnostic value.
T-Cell Large Granular Lymphoblastic Leukemia
T-Cell large granular lymphocytic leukemia (T-LGLL) is a chronic proliferation of clonal cytotoxic lymphocytes. The most common recurrent oncogenic mutations are STAT3 activating mutations, which are found in up to 70% cases and affect the SH2 domain of STAT3; while STAT5B SH2 domain mutations are less frequent and may be associated with aggressive disease. Mutations of these two genes can assist the diagnosis of T-LGLL and also serve as a signature of an aggressive clinical course and poor outcomes.10
Angioimmunoblastic T-cell Lymphoma
Angioimmunoblastic T-cell lymphoma (AITL) has frequent genetic alterations in epigenetic modifiers such as IDH2, TET2, and DNMT3A. TET2 and IDH1/2 mutations frequently coexist, even in the same cells, unlike myeloid malignancies, in which TET2 and IDH1/2 mutations are mutually exclusive.11
Anaplastic large cell lymphoma, ALK-positive
Anaplastic large cell lymphoma (ALCL), ALK-positive has the characteristic ALK fusion in 90% of cases. The most common partner gene is NPM1; while variant translocation partners include TPM3, ATIC, CLTC and others.12
Anaplastic large cell lymphoma, ALK-negative
Anaplastic large cell lymphoma (ALCL), ALK-negative are reported to have DUSP22 and TP63 alterations in approximately 30% and 8% cases. The 5-year overall survival rates are 90% for DUP22 while 17% for TP63 rearrangement.13 In addition, recurrent activating mutations of JAK1 and/or STAT3, which lead to constitutive activation of the JAK/STAT3 pathway, may have therapeutic implications in ALK-negative ALCL.14
Peripheral T cell lymphoma, not other specified type
Peripheral T cell lymphoma, not other specified type (PTCL-NOS) has complex pathogenesis.15 Recently, 2 subtypes of PTCL NOS, characterized by high expression of either GATA3 or TBX21, were identified as having prognostic and biologic significance.16
Summary
Advances in molecular testing technologies have changed the diagnosis, monitoring and disease management of T-cell malignances and may provide therapeutic targets. Understanding the basic principles of the molecular assays and the updated knowledge of genomic alterations will make practicing pathologists an integral part of patient management.
Listen to the podcast
Jinjuan Yao, MD, FCAP, member of the Personalized Health Care Committee explains how using next generation sequencing techniques allows more accurate initial diagnosis and more.
References
- Brüggemann M, Kotrová M, Knecht H, et al. Standardized next-generation sequencing of immunoglobulin and T-cell receptor gene recombinations for MRD marker identification in acute lymphoblastic leukaemia; a EuroClonality-NGS validation study. Leukemia. 2019;33(9):2241-2253.
- Ho CC, Tung JK, Zehnder JL, Zhang BM. Validation of a Next-Generation Sequencing-Based T-Cell Receptor Gamma Gene Rearrangement Diagnostic Assay: Transitioning from Capillary Electrophoresis to Next-Generation Sequencing. J Mol Diagn. 2021;23(7):805-815.
- Arcila ME, Yu W, Syed M, et al. Establishment of Immunoglobulin Heavy (IGH) Chain Clonality Testing by Next-Generation Sequencing for Routine Characterization of B-Cell and Plasma Cell Neoplasms. J Mol Diagn. 2019;21(2):330-342.
- Schumacher JA, Duncavage EJ, Mosbruger TL, Szankasi PM, Kelley TW. A Comparison of Deep Sequencing of TCRG Rearrangements vs Traditional Capillary Electrophoresis for Assessment of Clonality in T-Cell Lymphoproliferative Disorders. American Journal of Clinical Pathology. 2014;141(3):348-359.
- Marçais A, Lhermitte L, Artesi M, et al. Targeted deep sequencing reveals clonal and subclonal mutational signatures in Adult T-cell leukemia/lymphoma and defines an unfavorable indolent subtype. Leukemia. 2021;35(3):764-776.
- Choi S, Go JH, Kim EK, et al. Mutational Analysis of Extranodal NK/T-Cell Lymphoma Using Targeted Sequencing with a Comprehensive Cancer Panel. Genomics Inform. 2016;14(3):78-84.
- López-Nieva P, Fernández-Navarro P, Graña-Castro O, et al. Detection of novel fusion-transcripts by RNA-Seq in T-cell lymphoblastic lymphoma. Scientific Reports. 2019;9(1):5179.
- Kroeze E, Loeffen JLC, Poort VM, Meijerink JPP. T-cell lymphoblastic lymphoma and leukemia: different diseases from a common premalignant progenitor? Blood Adv. 2020;4(14):3466-3473.
- Zhang J, Ding L, Holmfeldt L, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481(7380):157-163.
- Teramo A, Barilà G, Calabretto G, et al. Insights Into Genetic Landscape of Large Granular Lymphocyte Leukemia. Front Oncol. 2020;10:152.
- Chiba S, Sakata-Yanagimoto M. Advances in understanding of angioimmunoblastic T-cell lymphoma. Leukemia. 2020;34(10):2592-2606.
- Zeng Y, Feldman AL. Genetics of anaplastic large cell lymphoma. Leuk Lymphoma. 2016;57(1):21-27.
- Parrilla Castellar ER, Jaffe ES, Said JW, et al. ALK-negative anaplastic large cell lymphoma is a genetically heterogeneous disease with widely disparate clinical outcomes. Blood. 2014;124(9):1473-1480.
- Pina-Oviedo S, Ortiz-Hidalgo C, Carballo-Zarate AA, Zarate-Osorno A. ALK-Negative Anaplastic Large Cell Lymphoma: Current Concepts and Molecular Pathogenesis of a Heterogeneous Group of Large T-Cell Lymphomas. Cancers (Basel). 2021;13(18).
- Xie C, Li X, Zeng H, Qian W. Molecular insights into pathogenesis and targeted therapy of peripheral T cell lymphoma. Experimental Hematology & Oncology. 2020;9(1):30.
- Zhang Y, Lee D, Brimer T, Hussaini M, Sokol L. Genomics of Peripheral T-Cell Lymphoma and Its Implications for Personalized Medicine. Front Oncol. 2020;10:898.

Jinjuan Yao, MD, PhD, FCAP, is a molecular pathologist from Memorial Sloan Kettering Cancer Center (MSK) with subspecialty training in molecular pathology and hematopathology. Dr. Yao also serves as the Quality Assurance Chair of Diagnostic Molecular Pathology at MSK.
Dr. Yao is a lifetime member of Chinese American Pathologists Association (CAPA) and the chair for CAPA molecular pathology subcommittee. She is also a CAP New York House of Delegates and CAP Personalized Health Care Committee (PHC) member.
Dr. Yao’s clinical work has focused on the large-scale, prospective genotyping of solid tumors and hematologic malignancies using a variety of NGS technologies. Her research interests include the pathogenesis of leukemias and lymphomas, genetic alterations, and therapies of solid tumors.