- Home
- Member Resources
- Articles
- Pharmacogenetics: A Tool for Precision Medical Therapy
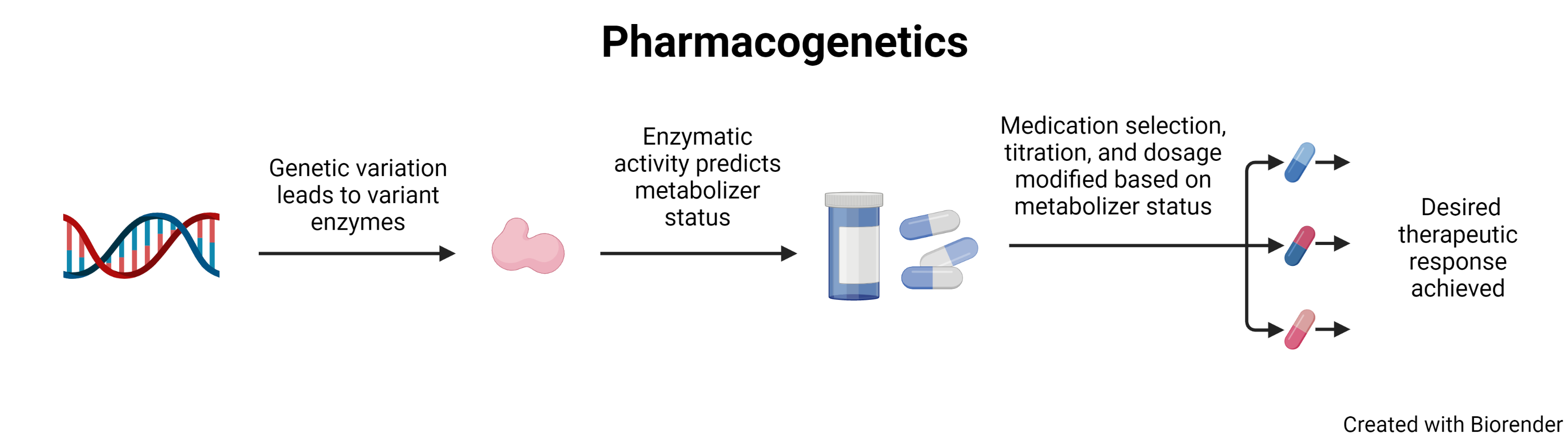
Pharmacogenetics, often used interchangeably with pharmacogenomicsi, is the study of how a patient’s genetic variation affects their response to medication. A patient’s genetics can predict how a patient will respond to drugs, while providers using this information can aim to increase efficacy and minimize toxicity. Implementing pharmacogenetics for patient care can be one avenue to bring precision medicine to patients by impacting medication selection, dosing, and titration.
The interaction between genes and medications was first discussed in medical literature of the 1950s,1 but the field has expanded rapidly recently as access to genetic information and computing has decreased in cost.2 Research has identified the genes important in the metabolism of hundreds of medications.3
Psychiatry is one area where pharmacogenetics has the potential to significantly impact patient outcomes. One landmark trial in this area of medicine is the Sequenced Treatment Alternatives to Relieve Depression (STAR*D), which implemented a stepwise approach to the treatment of major depressive disorder.4 STAR*D found that less than half of patients responded to the initial treatment of citalopram (Celexa). Of the nonresponders, between 1/4 and 1/3 also failed to respond to other selective serotonin reuptake inhibitors (SSRIs), serotonin and norepinephrine reuptake inhibitors (SNRIs), buproprion (Wellbutrin), or tricyclic antidepressants. In the trial, patients were on citalopram for up to 12 weeks, a length of time similar to the trial that many patients undergo when starting a new antidepressant drug. Relying on patient genetics, a clinician would be able to avoid treatment options unlikely to work and thereby remove a lengthy trial of a medicine ultimately likely to fail. Selecting the best medicine for the patient could lead to a more rapid reduction in clinical symptoms while avoiding adverse events associated with untreated mental health.
Identification of a patient’s genetic profile can be determined through several molecular assays, including polymerase chain reaction (PCR), Sanger sequencing, next-generation sequencing (NGS), or microarrays.
Patients have two copies of each gene linked to medication metabolism. To facilitate discussion of genetic variation, common variants are assigned an allele number. An allele is a defined sequence of a gene. The most common allele, also referred to as the ‘normal’ or the wild type, is defined as allele 1, represented by *1. Subsequent variants are assigned new allele numbers based on order of discovery. Therefore, the patient with wild type alleles can be represented as *1/*1.
In silico testing is performed on the protein created by the *1 allele and assigned an activity score. In silico testing refers to testing which relies on computer models to simulate the expected results for enzymes, as opposed to in vitro (testing performed in glass or test tubes) or in vivo (testing performed in living organisms) testing. The activity score established for the *1 allele is 1.0 Therefore, with two alleles, the wild type level of medication metabolism is 2.0. In silico testing can then be performed on variant alleles and, based on their ratio of metabolism to *1, assigned a score from 0 for non-functional alleles or above 1.0 for alleles that represent more rapid metabolism.
Combining the activity score of the patient’s two alleles, referred to as a diplotype, then gives a patient an overall activity score. This activity score places a patient in a metabolizer status category. The categories most used in literature are ultrarapid, rapid, normal, intermediate, or poor metabolizer.ii
Despite knowing the gene(s) linked to a specific medication’s metabolism, widespread implementation of pharmacogenetics has not occurred. One of the major issues has been how to interpret a patient’s metabolizer status. A busy clinician is unlikely to understand how being a poor metabolizer of a medication should impact its selection or dosage.
Seeking to mitigate this issue, a number of organizations around the world have been established to provide easy-to-follow recommendations to clinicians. These recommendations are generally based on the in silico testing as well as retrospectively studying responses to medications in patients with known genetic profiles. Three major organizations in this space include the Dutch Pharmacogenetics Working Group (DPWG), the Clinical Pharmacogenetics Implementation Consortium (CPIC), and the US Food and Drug Administration (FDA).
The Royal Dutch Pharmacist’s Association established the DPWG in 2005, and the organization publishes guidelines for gene-drug interactions. The DPWG guidelines are published with a scoring of evidence present and clinical relevance of the guidelines. The DPWG continues to update their guidelines.5
PharmGKB and the Pharmacogenomics Research Network started CPIC in 2009. CPIC has 26 guidelines published on its website for gene-drug interactions. CPIC guidelines are published with grading levels of evidence, how to assign phenotype to clinical genotypes, and prescription recommendations based on genotype. CPIC has active working groups that update recommendations.6
The FDA announced their publication of a three -section Table of Pharmacogenetic Associations (the “Table”) on February 20, 2020. Section one includes pharmacogenetic associations for which the data supports therapeutic management recommendations. Section two includes pharmacogenetic associations for which the data indicate a potential impact on safety or response, and section three includes pharmacogenetic associations for which the data demonstrate a potential impact on pharmacokinetic properties only. Despite the pandemic, the FDA has updated the Table multiple times since its launch.7
Guidelines provided by these organizations give clinicians an opportunity to practice precision medicine in medical therapies. However, some confusion exists because the organizations may provide different recommendations for the same medication, and in some cases, recommendations are highly divergent. A pathologist can function as a subject matter expert to clarify these recommendations for clinicians.
The lack of diversity in study populations is a significant weakness within the pharmacogenetic sphere. Many studies, and therefore recommendations, are based on data from populations of European descent.8 The prevalence of variants in different populations can be highly diverse. As more data becomes available in other populations, clinically significant alleles will be better understood. An example of this is CYP2C9 allele 8, *8. This was added to the Association of Molecular Pathologists (AMP) recommendations for pharmacogenetic testing after its prevalence was found to be significant within the African American community.9 Another example where this had profound legal consequences was in a recent lawsuit by the State of Hawaii against of clopidogrel (Plavix) producers, who knew that Asians and Pacific Islanders would be more likely to respond poorly to treatment which placed these patients at “grave risk of serious injury or death.”10 An $834 million award was upheld on appeal because the companies knew the different response, initially associated by ethnicity and later confirmed by genetics, to the medication could possibly be harmful.
Another issue facing pharmacogenetics is the lack of a single authority for purposes of standardization of results. Laboratories may be looking for different panels of alleles, and potentially a lab might report a patient as having wild type alleles, *1/*1, because their panel failed to identify an allele which another lab would identify. Recent efforts have included joint consensus statements on allele selection by AMP, the College of American Pathologists (CAP), DPWG, and the European Society of Pharmacogenomics and Personalized Therapy.11 A recent joint statement places clinically relevant alleles into one of two tiers. Tier 1 alleles “i) have a well-characterized effect on the function of the protein and/or gene expression, ii) have an appreciable minor allele frequency in a population/ethnicity group, and iii) have publicly available reference materials (RMs). Tier 2 alleles meet at least one of the three criteria for tier 1 alleles, but not all.” The consensus statement currently requests that at a minimum, laboratory testing includes tier 1 with tier 2 being an appropriate addition.
Despite the impediments that exist, many institutions are implementing adoption of pharmacogenetics based on the currently available data. Successful implementation of these programs should include pathologists due to their expertise in clinical medicine and the clinical laboratory. Pharmacogenetics will only continue to improve in utility as more data becomes available.
i. Pharmacogenetics is generally understood to refer to a single gene-drug interaction. Pharmacogenomics is generally understood to refer to the genome’s interaction with medications.
ii. Ultrarapid ≥ 3.0 Activity Score (AS), rapid > 2 but < 3 3 AS, Normal 2.0 AS, Intermediate 1.0 - 2.0 AS, Poor < 1.0 AS
References
- Kalow W, Staron N. On distribution and inheritance of atypical forms of human serum cholinesterase, as indicated by dibucaine numbers. Can J Biochem Physiol. 1957;35(12):1305-1320.
- National Human Genome Research Institute. The Cost of Sequencing a Human Genome. National Institute of Health. Updated November 1, 2021. Accessed April 1, 2023. https://www.genome.gov/about-genomics/fact-sheets/Sequencing-Human-Genome-cost
- Whirl-Carrillo M, McDonagh EM, Hebert JM, et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2012;92(4):414-417.
- Rush AJ, Trivedi MH, Wisniewski SR, et al. Acute and longer-term outcomes in depressed outpatients requiring one or several treatment steps: a STAR*D report. Am J Psychiatry. 2006;163(11):1905-1917.
- This is what the KNMP does. Royal Dutch Society for the Promotion of Pharmacy. Updated January 10, 2023. Accessed April 1, 2023. https://www.knmp.nl/index.php/over-de-knmp/dit-doet-de-knmp
- Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011;89(3):464-467.
- U.S. Food & Drug Administration. Table of Pharmacogenetic Associations. FDA. Updated October 26, 2022. Accessed April 1, 2023. https://www.fda.gov/medical-devices/precision-medicine/table-pharmacogenetic-associations
- Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature. 2016;538(7624):161-164.
- Pratt VM, Cavallari LH, Del Tredici AL, et al. Recommendations for clinical CYP2C9 genotyping allele selection: A Joint Recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn. 2019;21(5):746-755.
- Bellon T, Raymond N. Bristol-Myers, Sanofi ordered to pay Hawaii $834 million over Plavix warning label. Reuters. February 15, 2021. Accessed April 1, 2023. https://www.reuters.com/article/us-bristol-myers-sanofi-plavix/bristol-myers-sanofi-ordered-to-pay-hawaii-834-million-over-plavix-warning-label-idUSKBN2AF1YI
- Pratt VM, Cavallari LH, Del Tredici AL, et al. Recommendations for Clinical CYP2D6 Genotyping Allele Selection: A Joint Consensus Recommendation of the Association for Molecular Pathology, College of American Pathologists, Dutch Pharmacogenetics Working Group of the Royal Dutch Pharmacists Association, and the European Society for Pharmacogenomics and Personalized Therapy. J Mol Diagn. 2021;23(9):1047-1064.

Bradley Grant, MD, JD. The Human Genome Project sparked Dr. Grant’s interest in genetics, and he initially pursued this interest through law. Dr. Grant was an attorney in Washington state and admitted to practice before the United States Patent and Trademark Office before deciding to pursue medicine. As a resident, he is currently working to bring pharmacogenetic testing to patients at University of Texas Medical Branch Galveston. He is also interested in FDA regulatory policy and is involved in national organizations that sit at the intersection of law and medicine.

Jordan Laser, MD, is a board certified anatomic, clinical, and molecular genetic pathologist. Jordan is the Senior Director of Clinical and Medical Affairs at Bio-Techne as well as the CEO and Founder of Laser Laboratory Consulting. Previously, he served as Chief Laboratory Officer at Everly Health. The laboratory services at Everly Health support a wide range of consumer-initiated at-home tests. Having roots in New York, Jordan spent his first 11 years of practice as a pathology leader at Northwell Health. His expertise includes molecular and genomic medicine, laboratory management, healthcare finance, and standards and regulations.