- Home
- Member Resources
- Articles
- Molecular Subclassifications of DLBCL

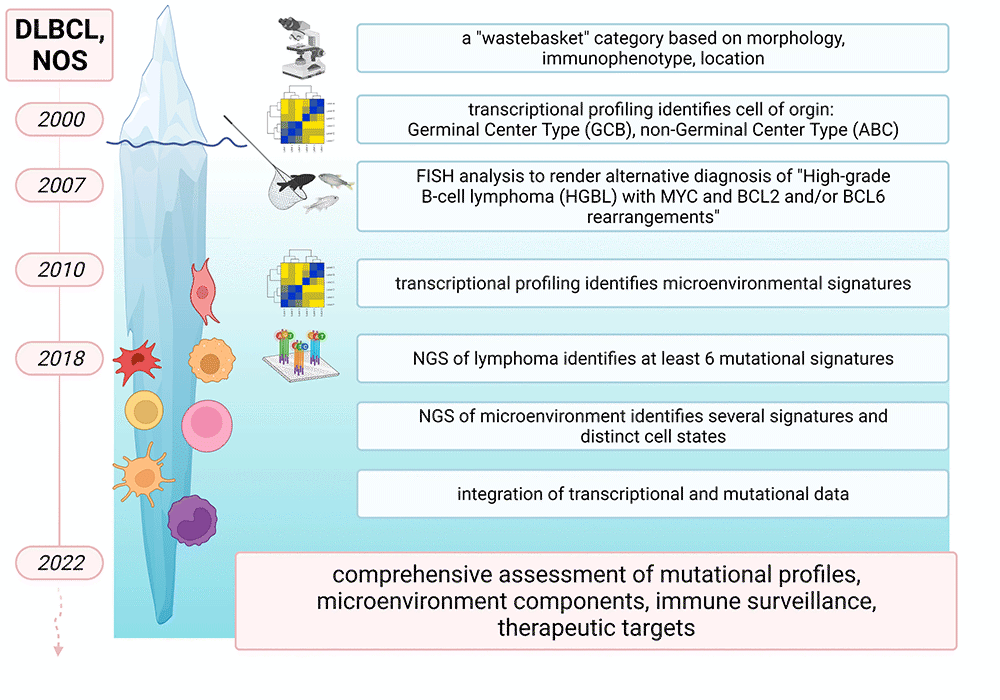
Figure 1: Iterative refinement of DLBCL-NOS and the lymphoma microenvironment by cytogenetic and molecular studies. Figure created with BioRender.com (under a full license)
Large B-cell lymphomas are a heterogeneous group of aggressive non-Hodgkin lymphomas (NHL). Treatment responses and patient outcomes vary greatly between subtypes and depend on whether lymphomas occur de novo or as transformation of a preexisting low-grade lymphoma, on anatomic location, or on morphologic variant.1 Most cases (up to 35% of NHL in Western countries) are classified as diffuse large B-cell lymphoma, not otherwise specified (DLBCL-NOS), as they do not fulfill morphologic, immunophenotypic or cytogenetic criteria for one of the specific subtypes or distinct entities according to the 2016 revision of the World Health Organization classification.2 Over the course of time, this "waste basket" category has undergone iterative prognostically relevant refinements, and it is now recognized that DLBCL-NOS exhibits structural rearrangements, complex copy number changes and somatic mutations that occur within intricate interactions with the non-malignant tumor microenvironment (TME).3,4 Herein we provide a high-level overview of how molecular studies have deconstructed DLBCL-NOS and its TME (Figure 1), and how these findings can improve diagnostic/ prognostic accuracy and identify treatment options (Figure 2).

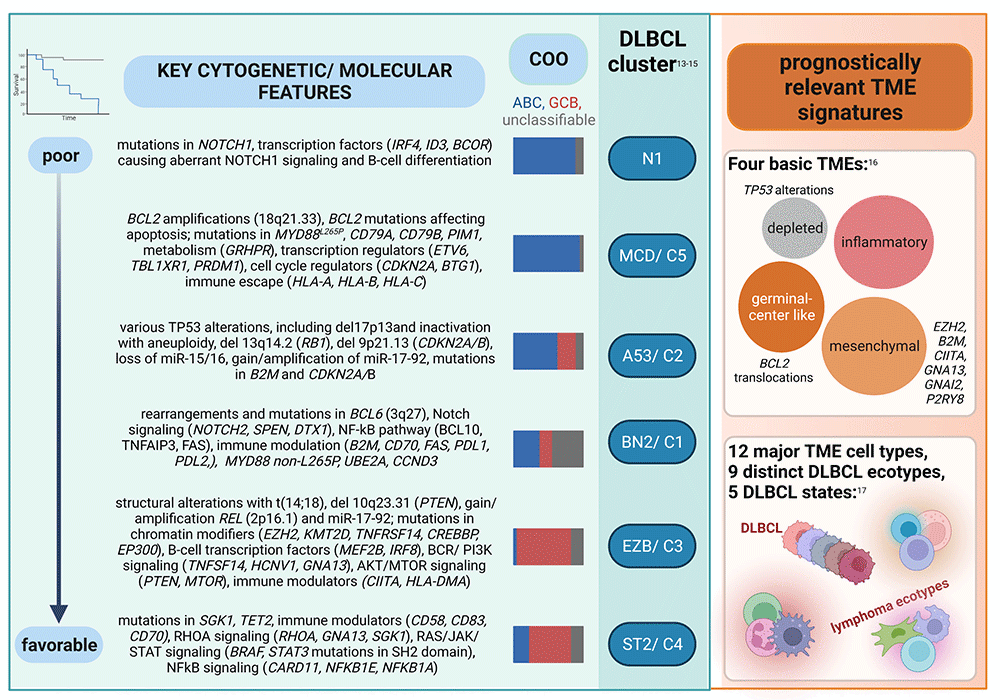
Figure 2: Somatic mutational and microenvironmental mutational signatures of DLBCL-NOS with biological and prognostic significance. Figure created with BioRender.com (under a full license)
Early microarray-based studies identified three prognostically relevant subtypes of DLBCL with gene expression patterns indicative of different stages of B-cell differentiation, also known as cell of origin (COO)5,6: a germinal center B-cell-like subtype (GCB-like), an activated B-cell-like (ABC-like) subtype, and a “type 3” category that did not cluster with either the GCB-like or ABC-like groups. As patients with GCB-like DLBCL have generally better overall survival than those with ABC-like DLBCL, both with and without rituximab-containing regimens, and better response to novel treatment approaches such as the Bruton's tyrosine kinase (BTK) inhibitor ibrutinib7, the 2016 WHO classification requires the identification of ABC-like versus GCB-like DLBCL.2 Additional gene-expression profiling (GEP) studies using a variety of methods, including GEP based on quantitation of RNA transcripts from formalin-fixed, paraffin-embedded tissue, further refined COO categories and provided additional insight into DLBCL pathogenesis, genomic composition and clinical behavior.4,8 To facilitate the assessment of COO in daily clinical practice, immunohistochemistry (IHC)-based surrogates have been developed. While not correlating perfectly with GEP, the Hans method using CD10, BCL6, and MUM1 protein expression is the most widely accepted classifier for GCB-like versus non-GCB-like determination.9 Compared to other methods such as the Choi (based on GCET1, CD10, BCL6, MUM1, and FOXP1 IHC)10, Visco (CD10, FOXP1, BCL611, or Tally algorithm (pairwise comparison of GCB [GCET1 and CD10] and ABC [FOXP1 and MUM1] immunophenotypes)12, Hans is considered the most “user-friendly” and most replicable, as it is based on only three commercially available antibodies that are commonly performed in most laboratories, yielding staining results that are relatively easy to interpret.
Subsequent to the COO categories, the prognostic significance of the combination of known recurrent structural rearrangements was illuminated. Currently, DLBCL-NOS must be distinguished from high-grade B-cell lymphomas, NOS, and from high-grade B-cell lymphomas with MYC and BCL2 and/ or BCL6 rearrangements ("double-hit" and/or "triple-hit" lymphomas). The later are associated with dismal outcome and poor response to immunochemotherapy and transplantation.2 As MYC, BCL2 and BCL6 rearrangements not meeting “double-hit"/ "triple-hit" diagnostic criteria are also present in 5%-15%, 20%-30% and 25%-45% of DLBCL, respectively, consensus guidelines recommend cytogenetic assessment (commonly performed as FISH studies using break-apart probes) for initial diagnostic workup.8 Of note, a subset of DLBCL-NOS referred to as “double-expressor” lymphomas exhibit MYC protein overexpression in the absence of cytogenetic evidence of MYC gene rearrangement, in association with BCL2 overexpression. Double-expressor DLBCL-NOS have inferior outcome compared to those lacking MYC/BCL2 overexpression, but are prognostically more favorable than bona fide "double-hit"/"triple-hit" lymphomas.8
With the advent of next generation sequencing (NGS) panel testing, prognostic categories of DLBCL based upon somatic mutational profiles were identified. Despite using fundamentally different algorithms, independent large-scale profiling efforts have identified robust, largely overlapping, and biologically and prognostically significant DLBCL signatures (Figure 2). Chapuy and colleagues at the Dana Farber Cancer Institute (DFCI) analyzed 304 primary DLBCL biopsies by combining different mathematical methods to cluster 158 genetic driver alterations, including recurrent mutations, somatic copy number alterations, and structural variants.13 This identified five clusters with distinct genetic signatures (C1–C5), and a cluster without any detectable alterations. Schmitz and colleagues at the National Institutes of Health (NIH) classified 574 DLBCL biopsies using the GenClass algorithm that starts with a set of genetic aberrations and then models all possible re-assortments into classes termed “seed categories” to optimize genetic distinctiveness.14 This approach identified four genetic subtypes (MCD, N1, BN2, and EZB). A subsequent GenClass algorithm-based study of over 1,200 DLBCL specimens discovered two additional genetic subclasses, A53 and ST2.15
Within the ABC-like subtype based upon COO, the worst prognosis is associated with the NIH N1 category (characterized by NOTCH1, ID3 and BCOR mutations). An unfavorable prognosis is also seen for the NIH MCD, which is highly concordant with DFCI Cluster C5. These DLBCLs are often associated with extranodal involvement and are enriched for concurrent MYD88 p.L265P and CD79B mutations, together with other mutations causing activated NF-kB signaling, increased proliferation, impaired apoptosis, and immune escape. An intermediate or more favorable prognosis is seen for BN2/Cluster 1 which shows a marginal zone-like mutation patterns characterized by BCL6 translocations, NOTCH2 mutations, and mutations in the B-cell receptor (BCR) and NF-κB signaling pathway. While the ABC-like subtype is still enriched in this category, there is a remarkable proportion of unclassifiable DLBCL. In contrast, EZB and Cluster 3 are almost exclusively composed of GCB-like DLBCL and are characterized by epigenetic and PI3K signaling pathway mutations and BCL2 translocations, as well as a variety of other genes involved in germinal center biology. ST2/Cluster 4, also predominantly composed of GCB-like DLBCL, is enriched for mutations affecting epigenetic regulation and PI3K and JAK/STAT signaling (nodular lymphocyte-predominant Hodgkin lymphoma [NLPHL]- and T cell histiocyte-rich large B cell lymphoma [THRLBCL]-like). Among the GCB-like subtype, ST2/Cluster 4 conveys a more favorable prognosis than EZB/Cluster 3. Finally, a separate category which overlaps both the ABC-like and GCB-like COO subtypes - A53/Cluster 2 - is characterized by various TP53 alterations, including inactivation with aneuploidy, and B2M and CDKN2A/B mutations. Together, these findings emphasized the genetic and prognostic diversity within GCB-like and ABC-like subgroups, and that constellations of mutations, structural rearrangements, and copy number alterations need to be considered.
Most recent large-scale molecular research has focused on gene and transcriptional signatures in the TME, and their integration with DLBCL mutational signatures. Kotlov and colleagues performed a transcriptomic analysis of the microenvironment of 4,655 DLBCLs and identified four basic prognostically and therapeutically relevant subtypes with varying associations with broad ABC-like/GCB-like categories and MCD/N1/A53/BN2/ST2 clusters - germinal center-like (higher proportions of follicular dendritic cells, endothelial cells, Tregs and T-helper cells), mesenchymal (enriched for vascular and endothelial cells and pathways including TGFB/SMAD and HIF), inflammatory (enriched in neutrophils, macrophages, CD8+ T-cells and PD-1+ CD8+ T cells), and depleted (scarce microenvironment).16 About one-third of TME germinal center-like cases harbored a BCL2 translocation, while other alterations (mutations in EZH2, B2M, CIITA, GNA13, GNAI2 and P2RY8) were enriched in the mesenchymal group. TP53 alterations and perturbation of cell cycle regulation was more common in DLBCL with a depleted TME subtype, which also had a higher proportion of A53 lymphomas. Steen and colleagues employed a machine-learning framework integrating transcriptome deconvolution and single-cell RNA sequencing from several large, independently collected datasets of bulk DLBCL transcriptomes to discover and validate DLBCL cell states and cellular ecosystems (“lymphoma ecotypes”).17 They identified 39 cell states across 12 different major TME cell types, nine distinct DLBCL cellular ecosystems, and five malignant B-cell states, and found survival associations of cell states enriched in ABC-like and GCB-like DLBCL, and of ABC and GCB cellular communities.
Accumulating evidence from clinical trials suggests that the adverse effects of certain genetic subtypes can be overcome by specific targeted treatment approaches. Wilson and colleagues demonstrated that adding ibrutinib to the standard rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) resulted in a 3-year 100% event-free survival in patients <60 years with MCD and N1 subtypes.18 Similarly, patients with unfavorable PIM1, SPEN or MYD88 mutations (associated with the BN2/Cluster 1 or MCD/Cluster 5 groups) or signatures including NF-κB, IRF4 and JAK-STAT (multiple subtypes including BN2/Cluster 1 and ST2/Cluster 4) benefited from the addition of the immunomodulatory drug lenalidomide to R-CHOP19, while patients with DLBCL with a distinct CD8 T-cell signature benefited from treatment with the proteasome inhibitor bortezomib and R-CHOP17. In contrast, the addition of venetoclax, a Bcl-2 inhibitor, to R-CHOP did not improve outcome of DLBCL patients with overexpression of BCL2 (EZB/Cluster 1 or MCD/Cluster 5), highlighting the complexity of DLBCL pathogenesis.20
Together, molecular studies have allowed a better understanding of the clinical heterogeneity and differences in responses to therapy in DLBCL-NOS. Future studies are needed to understand what DLBCL characteristics drive different TME compositions (and vice versa), to integrate molecular findings into established clinical prognostic and diagnostic algorithms, to develop clinically applicable surrogate markers, and to identify novel targeted treatment approaches. As we look to the future, a targeted knowledge-based biology-driven approach may point to more effective new treatment strategies, such as BLYM-777, a study designed to prospectively and comprehensively assess mutational profiles, TME components, immune surveillance and response to therapy.4
References
- Shanmugam V, Kim AS. Lymphomas. In: Tafe LJ, Arcila ME, eds. Genomic Medicine. Springer International Publishing; 2020:253-315.
- Swerdlow SH, Campo E, Harris NL, et al., eds. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Revised 4t. International Agency for Research on Cancer; 2017.
- Weber T, Schmitz R. Molecular Subgroups of Diffuse Large B Cell Lymphoma: Biology and Implications for Clinical Practice. Curr Oncol Rep. 2022;24(1):13-21.
- de Groot FA, de Groen RAL, van den Berg A, et al. Biological and Clinical Implications of Gene-Expression Profiling in Diffuse Large B-Cell Lymphoma: A Proposal for a Targeted BLYM-777 Consortium Panel as Part of a Multilayered Analytical Approach. Cancers. 2022;14(8):1857.
- Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503-511.
- Rosenwald A, Wright G, Chan WC, et al. The Use of Molecular Profiling to Predict Survival after Chemotherapy for Diffuse Large-B-Cell Lymphoma. N Engl J Med. 2002;346(25):1937-1947.
- Kuo HP, Ezell SA, Schweighofer KJ, et al. Combination of Ibrutinib and ABT-199 in Diffuse Large B-Cell Lymphoma and Follicular Lymphoma. Mol Cancer Ther. 2017;16(7):1246-1256.
- Kim AS, Wu CJ, Lovitch SB. Chapter 88: Molecular Genetic Aspects of Non-Hodgkin Lymphomas. In: Winthrobe’s Clinical Hematology. 14th ed. Lippincott Williams & Wilkins; 2019.
- Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103(1):275-282.
- Choi WWL, Weisenburger DD, Greiner TC, et al. A new immunostain algorithm classifies diffuse large B-cell lymphoma into molecular subtypes with high accuracy. Clin Cancer Res. 2009;15(17):5494-5502.
- Visco C, Li Y, Xu-Monette ZY, et al. Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lymphoma: a report from the International DLBCL Rituximab-CHOP Consortium Program Study. Leukemia. 2012;26(9):2103-2113.
- Meyer PN, Fu K, Greiner TC, et al. Immunohistochemical methods for predicting cell of origin and survival in patients with diffuse large B-cell lymphoma treated with rituximab. J Clin Oncol. 2011;29(2):200-207.
- Chapuy B, Stewart C, Dunford AJ, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018;24(5):679-690.
- Schmitz R, Wright GW, Huang DW, et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N Engl J Med. 2018;378(15):1396-1407.
- Wright GW, Huang DW, Phelan JD, et al. A Probabilistic Classification Tool for Genetic Subtypes of Diffuse Large B Cell Lymphoma with Therapeutic Implications. Cancer Cell. 2020;37(4):551-568.e14.
- Kotlov N, Bagaev A, Revuelta MV, et al. Clinical and Biological Subtypes of B-cell Lymphoma Revealed by Microenvironmental Signatures. Cancer Discovery. 2021;11(6):1468-1489.
- Steen CB, Luca BA, Esfahani MS, et al. The landscape of tumor cell states and ecosystems in diffuse large B cell lymphoma. Cancer Cell. 2021;39(10):1422-1437.e10.
- Wilson WH, Wright GW, Huang DW, et al. Effect of ibrutinib with R-CHOP chemotherapy in genetic subtypes of DLBCL. Cancer Cell. 2021;39(12):1643-1653.e3.
- Hartert KT, Wenzl K, Krull JE, et al. Targeting of inflammatory pathways with R2CHOP in high-risk DLBCL. Leukemia. 2021;35(2):522-533.
- Morschhauser F, Feugier P, Flinn IW, et al. A phase 2 study of venetoclax plus R-CHOP as first-line treatment for patients with diffuse large B-cell lymphoma. Blood. 2021;137(5):600-609.

Annette S. Kim, MD, PhD, FCAP is the Henry Clay Bryant Professor and Division Head of Diagnostic Genetics and Genomics at the University of Michigan. Dr. Kim’s research program has focused on the study of hematolymphoid malignancies, including miRNAs in myelodysplastic syndromes, myeloid and lymphoid mutational patterns, and test utilization management. She has served as a member of Molecular Oncology Committee and is currently Vice-Chair of the Personized Healthcare Committee for the College of American Pathologists. She is the Vice-Chair of the ASH Precision Medicine committee. In 2019, Dr. Kim was awarded the CAP Public Service Award.

Fabienne Lucas, MD, PhD, is a Fellow in Hematopathology at Brigham and Women's Hospital.