- Home
- Member Resources
- Articles
- Molecular Diagnosis and Targeted Treatment of Pancreatic and Biliary Cancers
Although pancreatic and biliary cancers are not ranked as the top five most common cancer types based on their incidence, they have very high mortality rates and are among the leading causes of cancer death in men and women combined in the United States. According to 2023 cancer statistics, there were an estimated 64,050 new cases of pancreatic ductal adenocarcinoma (PDAC) in 2023 and 52,550 estimated deaths. For gallbladder cancer and cholangiocarcinoma (CAA,) there were an estimated 12,220 new cases and 4,510 estimated deaths. Intrahepatic bile duct cancers, which are classified with liver cancer, had 41,210 estimated cases and 29,380 deaths.1 The high mortality rates of these cancers are largely related to their aggressiveness and the lack of effective treatment modalities. The overall five-year survival rate of all stages of pancreas and biliary cancer is only 10% - 15% with stage IV PDAC as low as 1%. It is essential to develop new diagnostic and treatment methods to detect these cancers at the early stage and treat them effectively to increase the survival. Molecular diagnosis and identification of predictive biomarkers of targeted treatment are integral to improving these patients’ outcomes.
The National Comprehensive Cancer Network (NCCN) guidelines recommend that both pan-cancer and tumor-specific biomarkers need to be tested. These predictive biomarkers have quite different incidence rates in these cancers, ranging from less than 1% to up to 30% (Table 1). Since they are associated with either approved or investigational therapeutic agents, it is critical to identify these alterations to provide potential survival benefits to the patients. The pan-cancer biomarkers are those approved by the Food and Drug Administration (FDA) in all cancer types with advanced or metastatic disease, including NTRK gene fusion, microsatellite instability high (MSI-H) or deficient mismatch repair (dMMR), and tumor mutational burden high (TMB-H). Cancer type-specific biomarkers associated with targeted therapy are seen at relatively high percentages in cholangiocarcinoma (CCA), with both FGFR2 fusion and IDH1 mutation approved by FDA in metastatic setting. CAA is divided into two subtypes: intrahepatic CCA (iCCA) and extrahepatic CCA (eCCA). IDH1/2 mutations are uncommon in eCCA. In iCCA, the mutation rates for IDH1 and IDH2 are 14% and 4%, respectively. IDH1 mutations are mainly R132C (69%) then R132L/G/S/H/F (total 31%), while IDH2 mutations occur at R172 (94.4%) and R140 (6.6%).2 Besides IDH1/2 and FGFR2, other genes commonly mutated in iCCA are BAP1 and TP53, while KRAS, SMAD4, and STK11 mutations are more common in eCCA.3
Table 1. Recommended predictive biomarkers in pancreas, biliary, and hepatocellular carcinomas
Recommended Biomarkers (yes or no) |
Cancer Types and Detection Rates (if available) |
||||
PDAC4 |
iCCA5 |
eCCA5 |
Gallbladder cancer5 |
Hepatocellular carcinoma6 |
|
NTRK 1/2/3 Gene fusion |
Yes |
Yes (< 1%) |
Yes (< 1%) |
Yes < 1% |
NA |
MSI-H/dMMR |
Yes |
Yes (1% - 3%) |
Yes (1% - 3%) |
Yes (1% - 3%) |
Yes |
TMB-H |
Yes |
Yes (< 5%) |
Yes (< 5%) |
Yes (< 5%) |
Yes |
BRAF V600E |
Yes |
Yes (1% - 5%) |
Yes (1% - 5%) |
Yes (1% - 5%) |
No |
FGFR2 fusion/ rearrangement |
No |
Yes (9% - 15%) |
Yes (Rare) |
No |
No |
IDH1 and IDH2 mutation |
No |
Yes (10% -20%) |
Yes (Rare) |
No |
No |
HER2 (ERBB2 overexpression and/or amplification) |
No |
Yes (5% - 20%) |
Yes (5% - 20%) |
Yes (15% - 30%) |
No |
RET gene fusion |
Yes |
Yes (< 1%) |
Yes (< 1%) |
Yes (< 1%) |
No |
BRCA1/BRCA2 |
Yes |
No |
No |
No |
No |
MSI-H: microsatellite instability-high; dMMR: mismatch repair deficient; TMB-H: tumor mutational burden-high |
|||||
Apart from the NCCN-guideline-recommended biomarkers, there are other important molecular findings in pancreatic and biliary cancers, such as alterations of KRAS, TP53, and CDKN2A genes. KRAS mutations, in particular, are present in up to 90% of PDAC. These mutations activate signaling pathways that promote cell proliferation and survival, contributing to tumor growth.7 The targeted treatment of KRAS G12D is under investigational study in PDAC.8 In KRAS negative cases, other driver mutations are commonly detected, including some druggable ones, such as fusions of FGFR2, NRG1, NTRK3, and ROS1, mutations of BRAF V600E, and ERBB2 amplification.9-10
Germline alterations are also important in PDAC, especially genes involved in DNA damage repair (DDR), since BRCA1/2 altered PDAC is sensitive to platinum-based therapy and poly (adenosine diphosphate–ribose) polymerase (PARP) inhibition with olaparib. Other DDR gene germline alterations, including ATM and PALB2, should also be tested in PDAC.11
Single gene testing using real time PCR (qPCR), dideoxy sequencing, fluorescence in situ hybridization (FISH),and immunohistochemistry (IHC), is still used in clinical labs. However, large panel next generation sequencing (NGS) assays are the most effective way to capture all types of genetic alterations and mutational signatures (especially with limited tissue), including single nucleotide variants (SNVs), insertions and deletions (Indels), copy number alterations (CNAs), structural variants (SVs), MSI-H, and TMB-H. Liquid biopsy is becoming more and more important in the molecular testing of cancers, both in the setting of initial diagnosis and in the disease monitoring post treatment. In comparison to tissue testing, it is less invasive, allows repeated analyses, and has a better representation of tumor heterogeneity and dynamic tumor progression. The limitation is mainly the low percentage of circulating tumor DNA (ctDNA), which may lead to false negative results, especially at the initial diagnosis.12
In addition to the guidance of targeted therapy, biomarker testing can also help with diagnosis and differential diagnosis, especially in the setting of cancer of unknown primary or in the distinction of primary or metastasis. These concepts are highlighted in the case discussed below.
A patient presented with a newly identified lung mass and a biopsy was performed. Based on morphology and TTF-1 immunohistochemistry, a diagnosis of lung adenocarcinoma was rendered (Figure 1, A and B). Further review of the medical history discovered that the patient had a prior diagnosis of CCA, which was stained weekly positive for TTF-1 (Figure 1-C). To distinguish the lung cancer as a separate primary or a metastatic lesion, both tumors were sequenced using a large panel NGS assay, which detected 4 shared mutations (listed in Table 2) with comparable variant allele frequencies (VAFs). Through the use of NGS testing of both cancers, the newly identified lung mass was diagnosed as a metastasis from the patient’s previous CCA, which was not possible based on morphology and TTF-1 staining alone. This finding dramatically effected this patient’s medical oncology management.
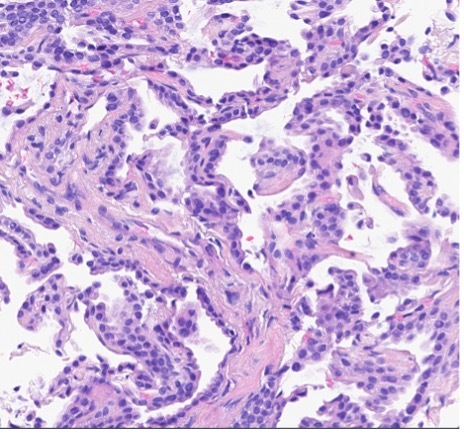
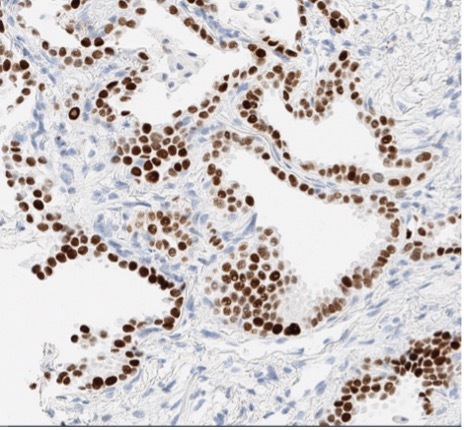
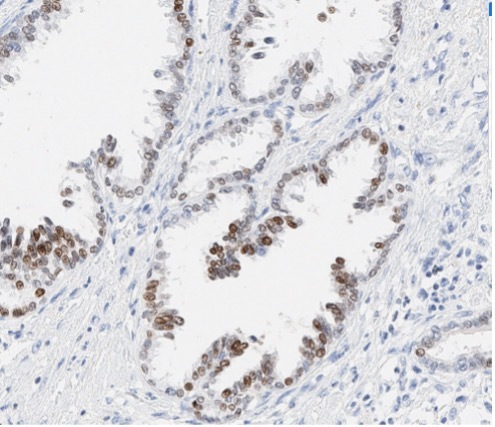
Figure 1. The H&E stain (A) of lung cancer and the TTF-1 IHC of both lung and biliary cancers (B and C). The morphology of lung cancer (A) is compatible with adenocarcinoma, which is strongly for TTF-1 IHC (B). The biliary cancer stained weakly positive for TTF-1 (C).
Table 2. The lung and biliary cancers shared alterations in this patient
Gene |
Alterations |
Biliary Cancer (VAF) |
Lung Cancer (VAF) |
||
TP53 |
p.R248L |
0.362 |
0.178 |
||
APC |
p.C2020Ffs*24 |
0.333 |
0.159 |
||
U2AF1 |
p.S34F |
0.271 |
0.130 |
||
ARID1A |
p.A898T |
0.241 |
0.154 |
||
VAF: Variant allele frequency |
|||||
To summarize, molecular testing of both pan-cancer and tumor specific biomarkers are essential for identification of targeted therapies of pancreatic and biliary cancers. The findings of genetic alterations can help pathologists in the establishment of diagnosis and/or differential diagnoses in morphologic and immunophenotypic challenging cases.
References
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17-48.
- Makawita S, Borad MJ, Carapeto F, et al. IDH1 and IDH2 Driven Intrahepatic Cholangiocarcinoma (IHCC): A comprehensive genomic and immune profiling study. J Clin Oncol. 2021;39:4009-4009.
- Lowery MA, Ptashkin R, Jordan E, et al. Comprehensive molecular profiling of intrahepatic and extrahepatic cholangiocarcinomas: potential targets for intervention. Clin Cancer Res. 2018;24(17):4154-4161.
- NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) Pancreatic Adenocarcinoma Version 2.2022 — December 6, 2022.
- NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) Biliary Tract Cancers Version 1.2023 — March 10, 2023.
- NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) Hepatocellular Carcinoma. Version 1.2023 — March 10, 2023.
- Cancer Genome Atlas Research Network. Electronic address: andrew_aguirre@dfci.harvard.edu, Cancer Genome Atlas Research Network. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017;32(2):185-203.e13.
- Chakravarty D, Gao J, Phillips SM, et al. Oncokb: a precision oncology knowledge base. JCO Precis Oncol. 2017;2017.
- Lowery MA, Jordan EJ, Basturk O, et al. Real-time genomic profiling of pancreatic ductal adenocarcinoma: potential actionability and correlation with clinical phenotype. Clin Cancer Res. 2017;23(20):6094-6100.
- Aguirre AJ. Oncogenic NRG1 fusions: A new hope for targeted therapy in pancreatic cancer. Clin Cancer Res. 2019;25(15):4589-4591.
- Javle M, Shacham-Shmueli E, Xiao L, et al. Olaparib monotherapy for previously treated pancreatic cancer with DNA damage repair genetic alterations other than germline BRCA variants: findings from 2 phase 2 nonrandomized clinical trials. JAMA Oncol. 2021;7(5):693-699.
- Zhao Y, Tang J, Jiang K, Liu SY, Aicher A, Heeschen C. Liquid biopsy in pancreatic cancer - Current perspective and future outlook. Biochim Biophys Acta Rev Cancer. 2023;1878(3):188868.

Jinjuan Yao, MD, PhD, FCAP, is a molecular pathologist from Memorial Sloan Kettering Cancer Center (MSK) with subspecialty training in molecular pathology and hematopathology. Dr. Yao also serves as the Quality Assurance Chair of Diagnostic Molecular Pathology at MSK.
Dr. Yao is a lifetime member of Chinese American Pathologists Association (CAPA) and the chair for CAPA molecular pathology subcommittee. She is also a CAP New York House of Delegates and CAP Personalized Health Care Committee (PHC) member.
Dr. Yao’s clinical work has focused on the large-scale, prospective genotyping of solid tumors and hematologic malignancies using a variety of NGS technologies. Her research interests include the pathogenesis of leukemias and lymphomas, genetic alterations, and therapies of solid tumors.