- Home
- Member Resources
- Articles
- Machine Learning in Pathology: The Potential to Predict the Future for Patients
Life with cancer is a perpetual state of high-stakes uncertainty. While those of us in healthcare derive some comfort from knowing the population-based data from clinical trials, when faced with an individual making treatment decisions that impact the quality of their often abruptly shortened expected lifespan, advice based on population data falls far short of satisfying. The incorporation of molecular pathology has provided significant insight into cancer pathogenesis, and even predictive capacity for a small subset of malignancies for which targeted therapies are available, but reliable predictions remain an elusive goal for the vast majority of patients. The inability to predict an individual’s course and/or best treatment likely contributes to the high prevalence of anxiety and depression among those of us living with cancer.1 Human costs aside, this likely also results in tremendous inefficiency in healthcare resource utilization. Pathologists may soon ameliorate both of these by incorporating the complementary strengths of machine learning into clinical practice to more thoroughly phenotype the pathophysiology and predict treatment response across numerous disease indications.
In machine learning (ML), a subset of artificial intelligence, algorithms are developed based on mathematical models that describe the distribution of known data; the algorithms then “learn” to make predictions or classifications on unknown data without explicit commands. There are two basic approaches, supervised and unsupervised learning. In supervised learning, human-derived annotations are used for training. In unsupervised learning, the model attempts to group un-annotated data to best fit some existing ground truth. Both of these approaches involve iterative attempts to refine the algorithm to best fit the known ‘ground truth’ that serves as input. Thus far, the majority of deep learning computer vision applications in pathology have relied on convolutional neural networks (CNNs), a supervised learning approach inspired by the architecture of the human visual cortex2 that uses pathologist-annotated histological images as input.3 CNNs have shown tremendous utility in analysis of digital whole slide images, and thus far most of the field has focused on pathological evaluation of cancer.4
Consider colorectal carcinoma (CRC), for which microsatellite instability (MSI) has implications for prognosis,5 therapeutic response,6,7 and the likelihood of an inherited cancer predisposition syndrome.8 Pathologists initially utilized manual evaluation of known characteristic morphologic features to identify patients for testing,9 but when data showed a relative lack of sensitivity using this approach,10 practice guidelines shifted to universal testing.11 Unfortunately, these techniques require additional resources that are not universally available.12 Algorithms with good performance characteristics could represent cost-effective screening tools. Can computers do better?
Preliminary data has been impressive: computers can be trained to identify MSI tumors across multiple cohorts on the basis of an H&E image alone, with sensitivity and specificity (AUCs ~0.89) similar to those seen with routine ancillary testing (using IHC and PCR).13,14 A CNN approach of has also been able to predict the gene expression-based consensus molecular subtype classification of CRC based on H&E images, provide additional measures of intratumoral spatial heterogeneity, and even propose a class for a subset of tumors that were unclassifiable by RNA expression profiling.15 ML applications have also shown promise in predicting other clinically relevant biomarkers, such as PD-L1 expression and clinically relevant molecular characteristics in lung non-small cell lung cancer.16,17
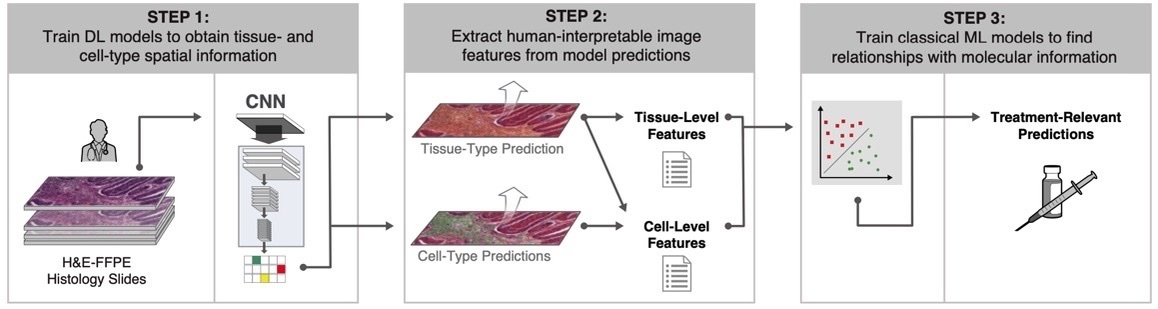
CNN models are often black box by nature. These approaches can sometimes show excellent performance, but with relative lack of explainability. One recent approach has combined the power of deep learning with interpretable features (termed human-interpretable features, or HIFs) (Figure 1).18 ML models were trained with >1.6 million pathologist annotations on >5,700 digitized whole H&E slide images representing five cancer types.

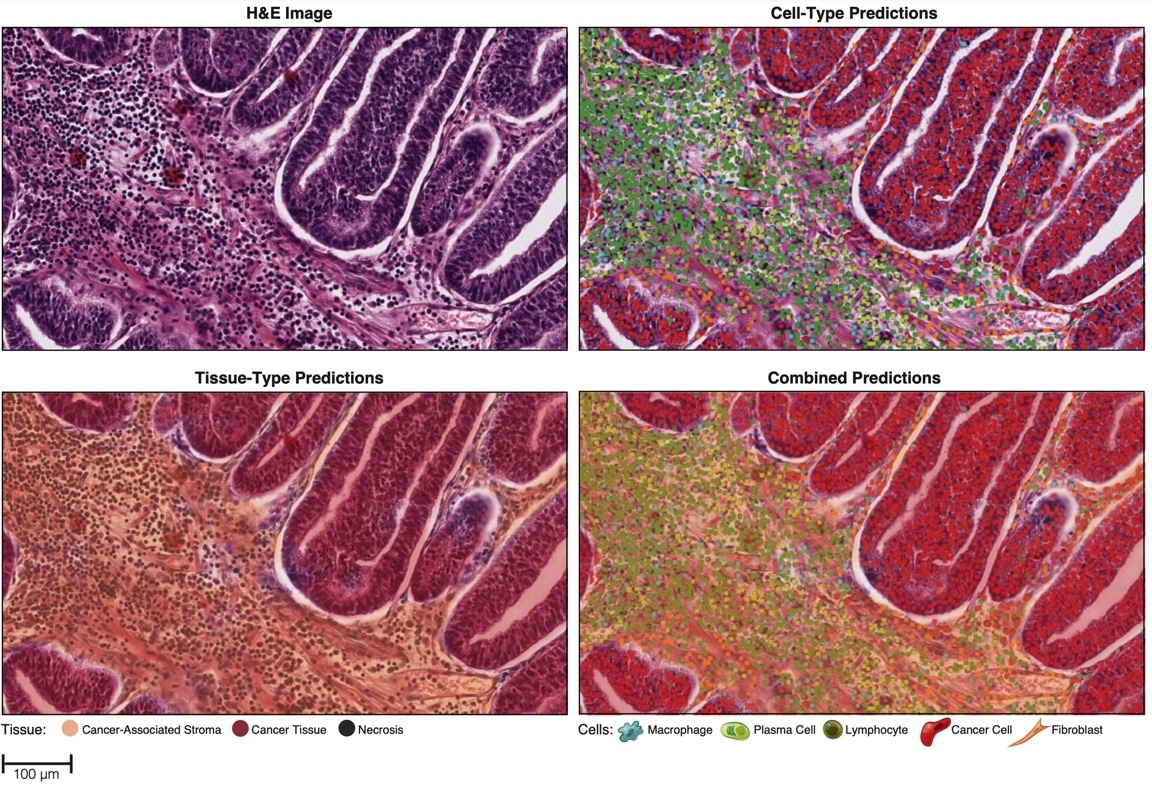
Figure 2. Representative images of cell and tissue predictions generated after model training.18
Model-generated cell- and tissue-type predictions (Figure 2) were then used to compute HIFs such as cell densities, inter-cellular spatial relationships, cell proximity features, and tissue area. The HIFs were associated with clinically relevant molecular phenotypes such as PD-1 and PD-L1 expression, homologous recombination deficiency score, and immune checkpoint receptor TIGIT expression. This HIF-based approach can enable rich feature extraction from the images to inform pathologist understanding of the features driving classification, facilitate hypothesis generation, and provide powerful insights into 'black box' models produced by CNNs.
One issue limiting the utility of CNNs is the dependence of their performance on the relationship between training data and the test set. Because AI is less flexible than humans when dealing with alterations in data quality and other preanalytic confounds, differences in patient characteristics, specimen type, tissue types, tissue fixatives, H&E stains, and image parameters can have detrimental effects on model performance, limiting the generalizability of the model. For instance, Echle et al. saw decreased performance when the algorithm trained on resection specimens was applied on biopsy specimens (AUC 0.96 vs 0.78). This is not surprising since the biopsies had less information, but also presumably included novel tissue types and procedural artifacts. Numerous approaches to increase the robustness of ML models are in development.
The above are just a few examples of proof-of-principle studies that provide encouraging evidence that AI-enhanced pathology brings clinically relevant added value to tissue-based analytics. Determining the best way to synthesize AI-assisted pathology with molecular and clinical pathology data to predict treatment responses will require incorporation of AI pathology tools into prospective clinical trials. This need is acknowledged by the recent creation of the FDA DDT program, whose scope includes utilization of AI-empowered surgical pathology to develop standardized biomarkers for clinical trials.
Because of the added complexity required to include clinical context to computer vision models, for the foreseeable future, these algorithms are best envisioned as a complementary assistive technology with vast potential to simultaneously enrich and refine pathology clinical diagnostics, translational research, and clinical trial results. The application of machine learning to pathology is still in its early stages, and there are many issues to solve19 before widespread deployment in clinical practice. However, given the recent FDA approval of slide scanners and algorithms for diagnostics, it seems the roadmap for development and regulation is becoming clear, and market forces are shifting toward cost-effectiveness,20 which ought to accelerate progress. The rate of progress and developments in AI is astonishing, making this an exciting time for pathology, as well as many other human endeavors. We look forward to pathology empowered by machine learning to extract quantitative insights, enabling precision pathology to ‘see the future’ using routine diagnostic specimens.
References
- Valentine, A. et al. Frequency of anxiety and depression and screening performance of the Edmonton Symptom Assessment Scale in a psycho-oncology clinic. Psychooncology (2021) doi:10.1002/pon.5813.
- LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
- Bera, K., Schalper, K. A., Rimm, D. L., Velcheti, V. & Madabhushi, A. Artificial intelligence in digital pathology - new tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 16, 703–715 (2019).
- Echle, A. et al. Deep learning in cancer pathology: a new generation of clinical biomarkers. Br. J. Cancer 124, 686–696 (2021).
- Sinicrope, F. A. et al. Prognostic impact of microsatellite instability and DNA ploidy in human colon carcinoma patients. Gastroenterology 131, 729–737 (2006).
- Sargent, D. J. et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J. Clin. Oncol. 28, 3219–3226 (2010).
- Ribic, C. M. et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N. Engl. J. Med. 349, 247–257 (2003).
- Cerretelli, G., Ager, A., Arends, M. J. & Frayling, I. M. Molecular pathology of Lynch syndrome. J. Pathol. 250, 518–531 (2020).
- Alexander, J. et al. Histopathological identification of colon cancer with microsatellite instability. Am. J. Pathol. 158, 527–535 (2001).
- Jenkins, M. A. et al. Pathology features in Bethesda guidelines predict colorectal cancer microsatellite instability: a population-based study. Gastroenterology 133, 48–56 (2007).
- Sepulveda, A. R. et al. Molecular biomarkers for the evaluation of colorectal cancer: Guideline from the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and American Society of Clinical Oncology. J. Mol. Diagn. 19, 187–225 (2017).
- Shaikh, T., Handorf, E. A., Meyer, J. E., Hall, M. J. & Esnaola, N. F. Mismatch Repair Deficiency Testing in Patients With Colorectal Cancer and Nonadherence to Testing Guidelines in Young Adults. JAMA Oncol 4, e173580 (2018).
- Kather, J. N. et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 25, 1054–1056 (2019).
- Echle, A. et al. Clinical-Grade Detection of Microsatellite Instability in Colorectal Tumors by Deep Learning. Gastroenterology 159, 1406–1416.e11 (2020).
- Sirinukunwattana, K. et al. Image-based consensus molecular subtype (imCMS) classification of colorectal cancer using deep learning. Gut 70, 544–554 (2021).
- Coudray, N. et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 24, 1559–1567 (2018).
- Sha, L. et al. Multi-Field-of-View Deep Learning Model Predicts Nonsmall Cell Lung Cancer Programmed Death-Ligand 1 Status from Whole-Slide Hematoxylin and Eosin Images. J. Pathol. Inform. 10, 24 (2019).
- Diao, J. A. et al. Human-interpretable image features derived from densely mapped cancer pathology slides predict diverse molecular phenotypes. Nat. Commun. 12, 1613 (2021).
- Cheng, J. Y., Abel, J. T., Balis, U. G. J., McClintock, D. S. & Pantanowitz, L. Challenges in the Development, Deployment, and Regulation of Artificial Intelligence in Anatomic Pathology. Am. J. Pathol. 191, 1684–1692 (2021).
- Lujan, G. et al. Dissecting the Business Case for Adoption and Implementation of Digital Pathology: A White Paper from the Digital Pathology Association. J. Pathol. Inform. 12, 17 (2021).
Authors
Michael Drage, MD, PhD, FCAP; Archit Khosla, MS; Mary Lin, PhD; Vicki Mountain, PhD; Eric Walk, MD, FCAP

Michael Drage is senior pathologist in the Scientific Programs Team at PathAI. He is a surgical pathologist with a background in immunology.

Eric Walk, MD, FCAP, is chief medical officer at PathAI and a member of the CAP Personalized Healthcare Committee. He has more than 20 years of experience in pathology, oncology drug development, precision medicine, and medical device development.