- Home
- Member Resources
- Articles
- Homologous Recombination Repair Deficiency (HRD): Brief Review of the Clinical Implication and Methodology of Measurement
Homologous recombination repair (HRR) is a high-fidelity process by which cells repair DNA double-strand breaks (DSBs) and is critical for maintaining genomic stability.1 Mutations or epigenetic alternations in any gene within the HRR pathway (such as BRCA1, BRCA2, ATM, BARD1, BRIP1, CDK12, CHEK1, CHEK2, FANCL, PALB2, RAD51B, RAD51C, RAD51D, and RAD54L) can result in HRR deficiency (HRD). HRD can lead to reliance on other error prone DNA repair pathways like non-homologous end joining (NHEJ) which causes genomic instability, leading to insertions or deletions in the nucleic acid sequences, abnormal copy number, and/or cross-linking of chromosomes.1
Clinical significance of HRD
HRD has been shown to be associated with sensitivity to platinum-based treatments and poly (ADP-ribose) polymerase inhibitors (PARPi). The aforementioned drugs induce cross-links between double-stranded DNA (dsDNA) that cancer cells with HRD cannot repair. PARPi work in concert with HRD through the mechanism of synthetic lethality (Figure 1).2 Poly (ADP-ribose) polymerase (PARP) plays a key role in DNA single-strand break (SSB) repair. PARPi inhibit the function of PARP and associated proteins by impeding dissociation of PARP from the DNA SSB leading to accumulation of SSBs and consequently DSBs. In healthy cells, the DSBs are eliminated by the mechanism of HRR. Cancer cells with HRD, however, are forced to use the error-prone NHEJ repair pathway leading to the gathering of genomic damage, which then results in apoptotic cell death. Consequently, PARP inhibition is particularly effective in cells with HRD through synthetic lethality (ie, combining two conditions, impairing both SSB and DSB repair, to force cell death, Figure 1).
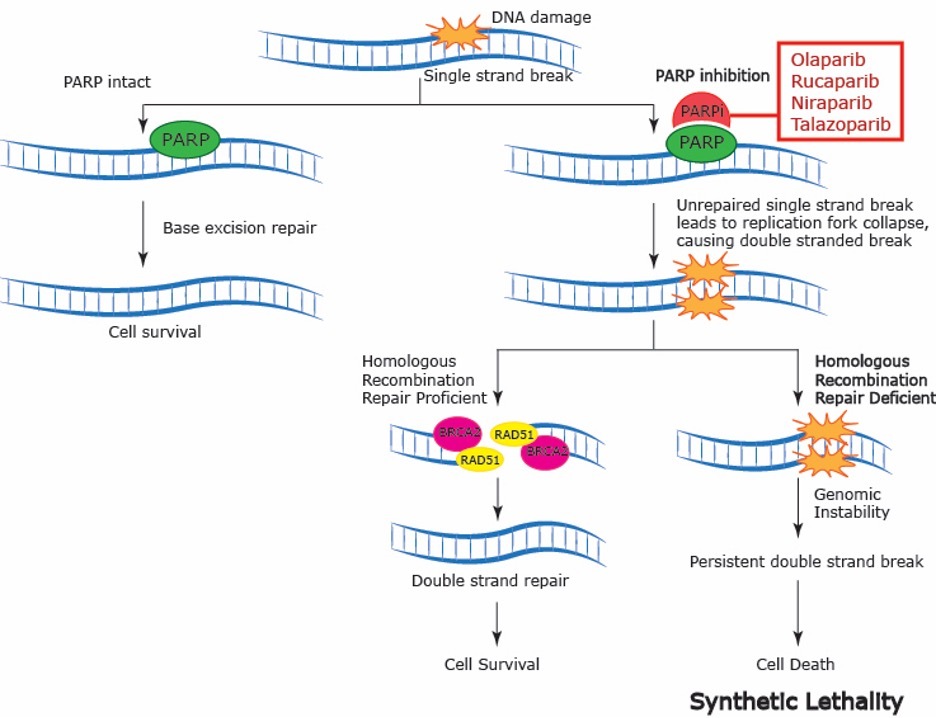
Summary of Treatment Implications
In the clinical landscape, changes have already been made to the standard of care (eg, National Comprehensive Cancer Network [NCCN] guidelines) based on a patient’s BRCA1/2 mutation and/or HRD status (Table 1: HRD as a Biomarker for PARP Inhibitors). Current NCCN guidelines for the treatment of patients with epithelial ovarian cancer recommends platinum-based therapy as primary adjuvant therapy. Patients who have complete or partial response to primary therapy, in the presence of either germline or somatic BRCA1/2 mutation, and have not been exposed to bevacizumab are strongly recommended (category 1) to add PARPi (olaparib or niraparib) as maintenance therapy.3 In patients who have been exposed to bevacizumab in primary therapy, in the presence of germline or somatic BRCA1/2 mutation and/or HRD phenotype as determined by tests described below, maintenance therapy with olaparib in combination with bevacizumab is strongly recommended (category 1).3 However, patients who are BRCA1/2 wild-type, HRR proficient, or unknown are recommended to continue with bevacizumab alone as maintenance.3
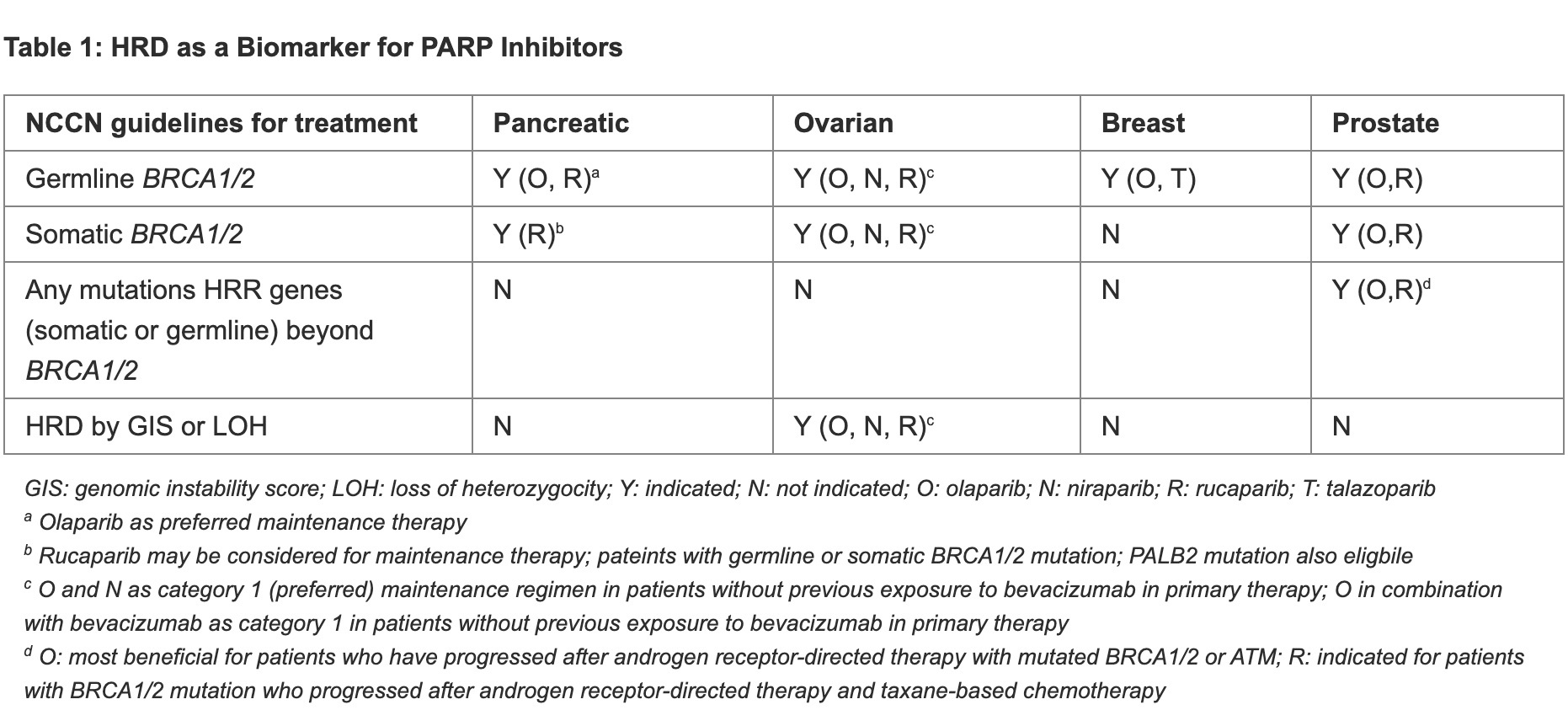
In HR-positive and HER2-negative breast cancer patients with germline BRCA1/2 mutation, PARP inhibition with olaparib or talazoparib is the recommended first line therapy for recurrent unresectable or metastatic diseases.4 In addition, patients who have triple negative breast cancer (TNBC) with germline BRCA1/2 mutation and are PD-L1 negative are recommended either PARPi (olaparib or talazoparib) or platinum-based chemotherapy (cisplatin or carboplatin) as the first line therapy for recurrent unresectable or metastatic diseases.4
For patients with pancreatic cancers, gemcitabine + cisplatin as initial systemic therapy is one recommended regimen that is only indicated in patients with BRCA1/2 or PALB2 mutations.5
Furthermore, for patients who have metastatic disease and have previously undergone platinum-based chemotherapy, olaparib is the recommended maintenance therapy for those having germline BRCA mutations; while rucaparib may be considered for patients with germline or somatic BRCA1/2 or PALB2 mutations.5
In prostate cancer, olaparib treatment may be considered for patients with mutations (germline or somatic) in any of the HRR pathway genes who have disease progression after androgen-receptor directed treatment. Efficacy with olaparib, however, is mainly observed in patients who have at least one mutation in the BRCA2, BRCA1, or ATM genes, particularly in patients with BRCA1/2 mutations.6 Patients who are BRCA1/2 mutated (germline or somatic) and have not been exposed to hormone therapy may be treated with olaparib in combination with abiraterone. In addition, rucaparib can be used in patients with BRCA1/2 mutations (germline or somatic) who have received androgen deprivation therapy and taxane-based chemotherapy.6
Measurement of HRD status
The measurement of HRD remains, however, challenging in clinical practice. A positive HRD status can be defined by the presence of pathogenic or likely pathogenic mutations in HRR genes (eg, BRCA1/2) and/or genomic instability caused by deficiency of HRR pathway. Most of HRD cases caused by mutations (single nucleotide variants and small insertions/deletions) in the genes involving HRR pathway can be readily detected by the current short-read based next-generation sequencing (NGS)-based assays (either whole genome/exosome or comprehensive targeted gene panels) commonly used in the laboratories. However, HRD cases caused by epigenetic silencing or large deletion may be missed by these assays.
Instead, several methods have been developed to detect these cases by using the hallmark of HRD phenotype, represented by genomic instability, which can be visible as genomic scars using either NGS-based or single nucleotide polymorphism (SNP) array-based methods. Measures of genomic scarring include loss of heterozygosity (LOH), telomeric allelic imbalance (TAI), and large-scale state transitions (LST).1,7,8 Measurement of HRD phenotype through these genomic scars enables the identification of significant percentages of patients with HRD status but without detectable mutations in HRR pathway genes. For example, the data from the trial (NCT02655016) evaluating frontline maintenance niraparib in patients with advanced ovarian cancers showed about 20% of patients exhibited the HRD phenotype without BRCA1/2 mutations when measured by genomic instability sore (GIS) using Myriad myChoice CDx (see the following paragraphs).7 Of note, the patients with HRD phenotype (i,. GIS ≥ 42) without BRCA1/2 mutations had survival data similar to the patients with mutated BRCA1/2.7
SNP-based platforms are capable of measuring copy number variants (CNVs) and LOH to estimate genomic scars. Two commonly used SNP arrays are OncoScan Dx (Affymetrix) and Infinium CytoSNP-850K BreadChip v1.2 (Illumina) capable of determining LOH, LST, and TAI.7,8 However, these methods cannot detect mutations of genes involving HRR pathway.
NGS based methods, besides detecting mutations of HRR pathway genes, are also capable of measuring genomic scars when used in combination with appropriate bioinformatics algorithms.7,8 Currently, the FDA has approved two companion diagnostic assays on the basis of their detection of the HRD phenotype. The Myriad myChoice CDx, available from Myriad, is the companion diagnostic for niraparib for use in patients with epithelial ovarian cancer. This test used DNA isolated from formalin-fixed paraffin embedded (FFPE) tumor tissue specimens to assesses the GIS, which measures LOH, TAI, and LST. HRD phenotype is considered positive when GIS is ≥ 42.1,7 Of note, this CDx can also detect the mutations in BRCA1/2 beyond HRD phenotype.
The FoundationFocus CDxBRCA assay (Foundation Medicine) is approved as the companion diagnostic for rucaparib. This assay uses an LOH score to define the presence of an HRD phenotype with an LOH score ≥ 16. In the clinical trial setting (ARIEL3 trial, NCT01968213), HRD was defined by LOH score ≥ 16 and/or mutations of BRCA1/2. Of note, in this trial, the HRD phenotype was identified in about 28% of patients without detectable BRCA1/2 mutations7. This test is also performed as part of the FoundationOne CDx, which is capable of detecting mutations of HRR pathway genes (including BRCA1/2) as well as quantifying LOH.1,7,8,9 There are additional large reference laboratories having developed their own HRD phenotype measuring assays, such as HRD genomic scar score by Caris Life Sciences. However, the majority of these assays have not been tested in the setting of clinical trials, and the clinical validity of these assays remains unproven.
Besides the above central laboratory-based assays for measuring HRD phenotype, several commercially available research only assays have recently become available to laboratories for internal validation to use as a laboratory developed assays to evaluate HRD phenotype. Examples of these assays include TruSight Oncology 500 HRD (Illumina), Oncomine Comprehensive Assay Plus (Thermo Fisher Scientific), and Sophia DDM HRD Solution (Sophia Genetics). The Illumina assay has licensed the GIS from Myriad. The Oncomine assay uses a genomic instability metric (GIM), and the Sophia assay uses genomic integrity index (GII) to determine the HRD phenotype. GIM is based on genomic segmentation using CNV log2 ratios and log odds for SNP allele frequencies, allowing the measurement of different unbalanced copy number events across autosomes. GIM ranges from 0 - 100, and higher values indicate more genomic instability with a cutoff of ≥ 16 defining HRD phenotype.10 GII uses a deep-learning based solution using low-pass whole genome sequencing (lpWGS) data (x1 fold coverage) to measure the genomic scar.11 Of note, lpWBS may be not reliable in measuring LOH, which needs accurate measurement of variant allele fractions determined by significant depth of coverage of SNP loci.12 Nevertheless, GII shows good agreement with GIS by Myriad assay.11 With individual assays, however, cutoffs for determining HRD phenotype are different due to different methodologies used. It is important to comprehensively evaluate the clinical utilities of measuring HRD phenotypes by these proprietary assays.
It is important to note that genomic scars represent the past genomic changes caused by HRD and may not reflect the current status of HRR efficacy.1 For example, studies have revealed reversion of BRCA mutation may occur in cancer cells leading to HRR proficiency after occurrence of genomic scars.13 This may contribute to the resistance of PARPi in patients with high GIS or high LOH by current testing methods.
In summary, HRD status is an important biomarker for selecting appropriate therapies (PARPi and/or platinum-based chemotherapy) for patients with ovarian, breast, pancreas, and prostate cancers. There is an urgent need for the clinical laboratories to develop methods to accurately evaluate HRD status and to harmonize various assays currently in uses.
References
- Stewart MD, Merino Vega D, Arend RC, et al. Homologous recombination deficiency: concepts, definitions, and assays. Oncologist. 2022;27(3):167-174. doi:10.1093/oncolo/oyab053
- Wang R, Han Y, Zhao Z, et al. Link synthetic lethality to drug sensitivity of cancer cell. Brief Bioinform. 2019;20(4):1295-1307. doi:10.1093/bib/bbx172
- National Comprehensive Cancer Network. NCCN Guidelines - Ovarian Cancer. Version 1.2024. Accessed January 26, 2024. https://www.nccn.org/professionals/physician_gls/pdf/ovarian.pdf
- National Comprehensive Cancer Network. NCCN Guidelines - Breast Cancer. Version 1.2024. Accessed January 26, 2024. https://www.nccn.org/professionals/physician_gls/pdf/breast.pdf
- National Comprehensive Cancer Network. NCCN Guidelines - Pancreatic Cancer. Version 1.2024. Accessed January 26, 2024. https://www.nccn.org/professionals/physician_gls/pdf/pancreatic.pdf
- National Comprehensive Cancer Network. NCCN Guidelines - Prostate Cancer. Version 1.2024. Accessed January 26, 2024. https://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf
- Ngoi NYL, Tan DSP. The role of homologous recombination deficiency testing in ovarian cancer and its clinical implications: do we need it? ESMO Open. 2021;6(3):100144. doi:10.1016/j.esmoop.2021.100144. Epub 2021 May 18.
- Wagener-Ryczek S, Merkelbach-Bruse S, Siemanowski J. Biomarkers for homologous recombination deficiency in cancer. J Pers Med. 2021;11(7):612. doi:10.3390/jpm11070612
- Ford L, Wolford JE, Brown SM, Randall LM. A profile on the FoundationFocus CDxBRCA tests. Expert Rev Mol Diagn. 2020;20(3):285-292. doi:10.1080/14737159.2020.1701438
- Dumur CI, Krishnan R, Almenara JA, et al. Analytical validation and clinical utilization of the Oncomine Comprehensive Assay Plus Panel for comprehensive genomic profiling in solid tumors. J Mol Pathol. 2023;4(2):109-127. doi:10.3390/jmp4020012
- Pozzorini C, Andre G, Coletta T, et al. GIInger predicts homologous recombination deficiency and patient response to PARPi treatment from shallow genomic profiles. Cell Rep Med. 2023;4(12):101344. doi:10.1016/j.xcrm.2023.101344
- de Luca XM, Newell F, Kazakoff SH, et al. Using whole-genome sequencing data to derive the homologous recombination deficiency scores. NPJ Breast Cancer. 2020;6:33. doi:10.1038/s41523-020-0172-0
- Pettitt SJ, Frankum JR, Punta M, et al. Clinical BRCA1/2 reversion analysis identifies hotspot mutations and predicted neoantigens associated with therapy resistance. Cancer Discov. 2020;10(10):1475-1488. doi:10.1158/2159-8290.CD-19-1485

Katherine Gui is currently a second-year medical student at Florida State University (FSU) College of Medicine. She earned her bachelor of health science degree at the University of Florida. In conjunction with her studies, Ms. Gui has dedicated her time advocating on behalf of medical students in the Medical Student Council as a representative in the FSU Congress of Graduate Students and as member of the Medical Student Wellness Committee. Her areas of interests include women’s health and equity and access in medicine. She is excited to enter her third-year rotations to continue her work as an advocate for her patients.

Chung-Che (Jeff) Chang, MD, PhD, FCAP, is the medical director of Hematology and Molecular/Genomic Laboratory at AdventHealth-Orlando, and professor of pathology, University of Central Florida. He currently serves as associate editor for Archives of Pathology and Laboratory Medicine, and a member of the Personalized Healthcare Committee for the College of American Pathologists (CAP). He was the principal investigator of several National Institutes of Health (NIH)/National Cancer Institute (NCI) grants to study myeloma, myelodysplastic syndromes, and lymphoma. Dr. Chang’s research interests include the use of next-generation sequencing technologies for clinical diagnostics and biomarker discovery.