- Home
- Member Resources
- Articles
- An Emerging Role of Pathologists in KRAS Mutation Testing and Targeted Treatment
KRAS is the most common oncogenic driver gene in human cancers, with the highest mutation frequency in pancreatic adenocarcinoma, followed by colorectal adenocarcinoma (CRC), cholangiocarcinoma, and non-small cell lung cancer (NSCLC) (Table 1, Figure 1).
Table 1. KRAS mutation frequency across cancer types
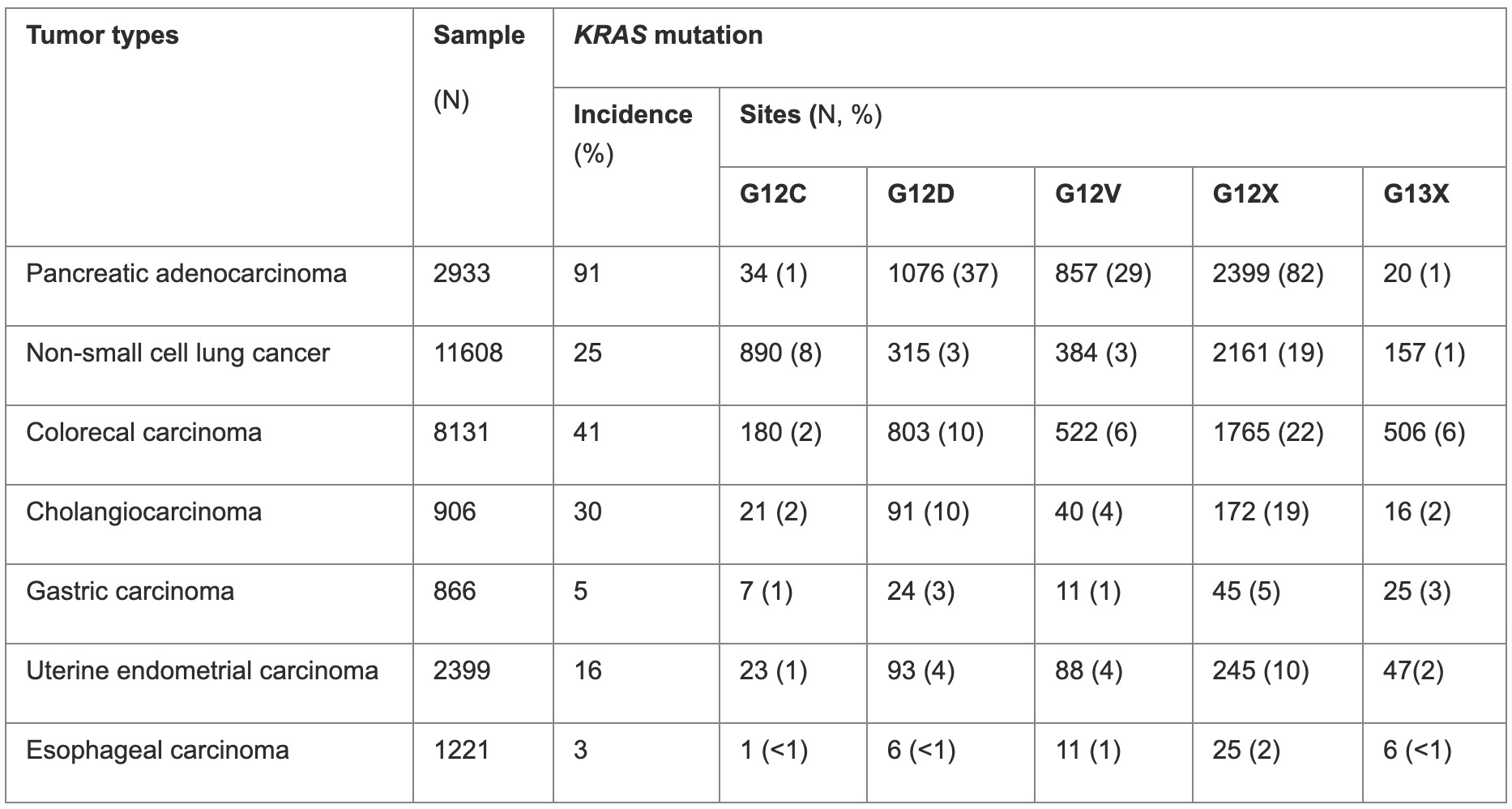
Data extracted using the publicly available cases from www.cBioPortal.org
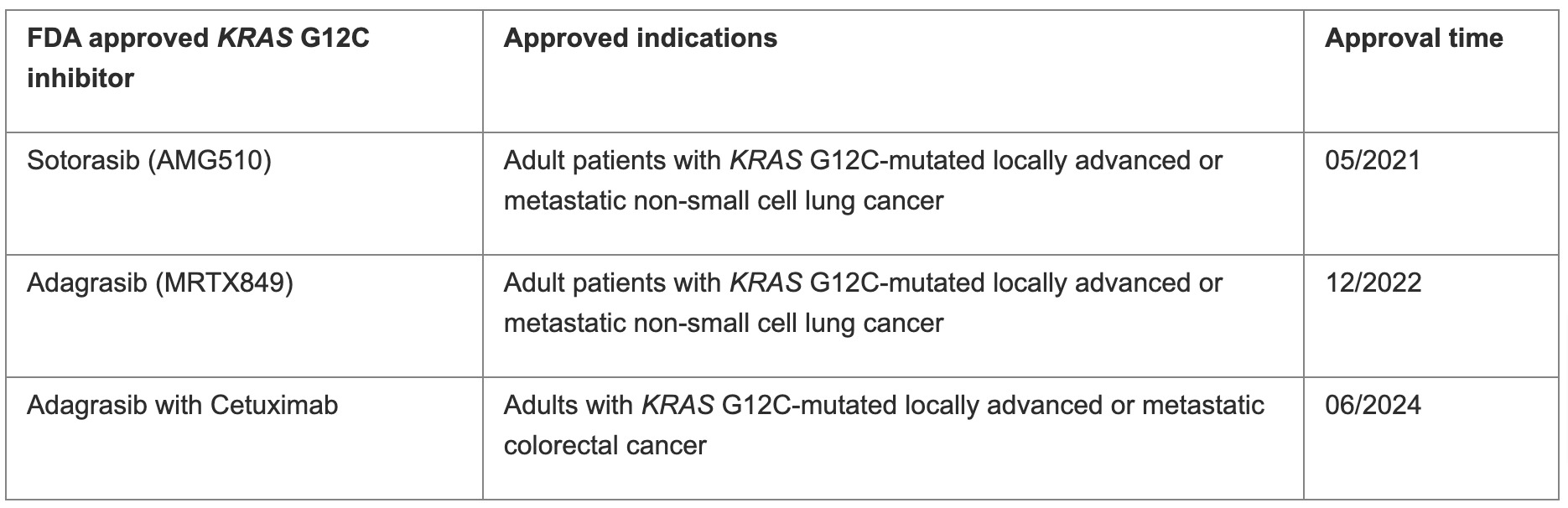
Because of the high prevalence of KRAS mutations across cancer types, targeting KRAS is an appealing strategy. In recent years, notable progress has been made, particularly in targeting KRAS (G12C). A class of covalently specific small molecules, including sotorasib (AMG510) and adagrasib (MRTX849), were developed and demonstrated promising results in clinical trials. These drugs have been approved by the FDA for adult patients with KRAS G12C mutated locally advanced or metastatic NSCLC, in 2021 and 2022, respectively. In addition, FDA also approved adagrasib plus cetuximab for adults with KRAS G12C mutated locally advanced or metastatic CRC (Table 2).3 Inhibitors targeting other KRAS mutations are also in development and early clinical trials. Therefore, accurately detecting KRAS mutations is crucial for selecting patients who may benefit from these targeted therapies.
Table 2. FDA-approved KRAS G12C inhibitors
This article will discuss the frequency of KRAS mutations in various cancer types, introduce commonly used testing methods, and explore available KRAS inhibitors. Additionally, it will focus on the current challenges and emerging role of pathologists in this evolving field.
KRAS mutations frequency and biochemical heterogeneity
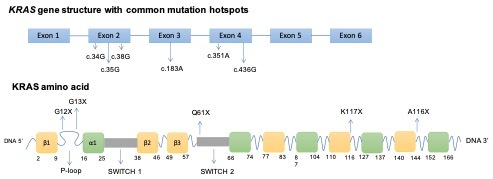
KRAS mutations vary between different cancer types (Table 1). The vast majority of KRAS mutations are single-base missense mutations, commonly found at codons 12 (83%), 13 (14%), or 61 (2%) on exons 2 and 3 (Figure 2).4
In pancreatic adenocarcinoma, KRAS
G12D and G12V are the predominant mutations, with a frequency of 40% and 32%, respectively. In lung adenocarcinoma, G12C is the most common among smokers (44%), followed by G12V (19%). G12D is the most frequent mutation in never-smokers (56%).5
Right-sided colorectal tumors have a predilection of KRAS mutations and G12 mutations account for up to 65%. Similar to pancreatic cancer, G12D and G12V are dominant mutation subtypes. Notably, G13 mutations make up about 18% of all KRAS mutations in colon cancers but are almost never found in pancreatic and lung cancers. Mutations at codons 117 and 146 are seen exclusively in colon cancers.6
Interestingly, KRAS mutations exhibit biochemical diversity. G12 and G13 mutations, located on the P loop, affect nucleotide stabilization and hydrolysis, with G12C retaining some normal guanosine triphosphate (GTPase) activity.5 Codon 13 mutations also increase nucleotide exchange.7 Q61 mutations, at the N-terminus of switch II, enhance guanosine diphosphate (GDP)-guanosine triphosphate (GTP) exchange but inhibit GTP hydrolysis, having the lowest hydrolysis rate among KRAS mutations.7 Different KRAS mutations activate specific signaling pathways. G12A, G13D, and Q61L activate the MAPK pathway, G12C and G12V activate RAL signaling, and G12D activates the PI3K-AKT-mTOR pathway.8
Detection of KRAS mutations
KRAS mutations can be detected by many different methods, including Sanger sequencing, real-time PCR (RT-PCR), high-resolution melting (HRM) analysis, mutant-enrichment PCR, such as co-amplification at lower denaturation temperature-PCR (COLD-PCR) and peptide nucleic acid (PNA)-clamp assay, pyrosequencing, as well as next generation sequencing (NGS).9 The characteristics of these methods are summarized in Table 3.
Table 3. Summarization of KRAS testing methods.
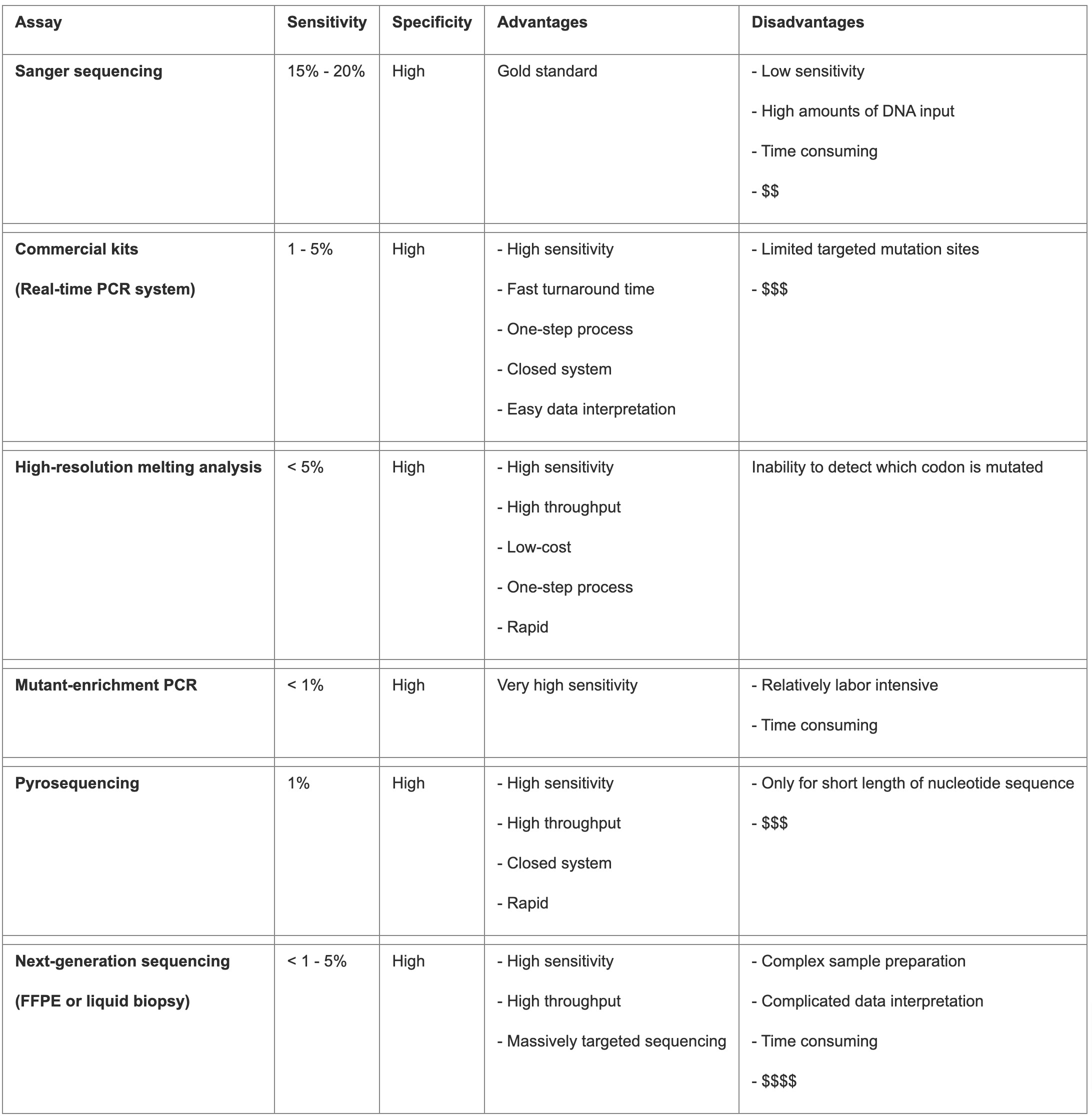
Direct sequencing, particularly PCR followed by dideoxy sequencing, was the gold standard for mutation detection but requires a high mutation allele frequency (10-30%), limiting its sensitivity.9 Other methods with higher sensitivity while still comparable specificity have been developed and are utilized clinically, either single gene or NGS-based. The TheraScreen KRAS kit (Qiagen), utilizing ARMS technology, is FDA-approved, detecting seven KRAS mutations with higher sensitivity and specificity than direct sequencing.10 The StripAssay (Vienna Labs) and SNaPshot techniques detect additional mutations with varying sensitivity, cost, and flexibility.10 The Cobas TaqMelt PCR assay (Roche) detects 19 KRAS mutations, offering high reproducibility and rapid results. However, these methods cannot detect rare mutations, limiting their clinical utility.11 NGS-based testing provides comprehensive mutation analysis, including all clinically relevant KRAS mutations, along with other genetic alterations and mutational signatures, and is widely used despite its higher cost.
Accurate detection is the prerequisite for guiding clinical decision-making. When implementing a KRAS mutation testing platform in a clinical setting, it is important for surgical and molecular pathologists to consider several factors, such as available specimen (tissue biopsy, resection, cytology, or liquid biopsy), space, instruments, labor, cost-effectiveness, expected sensitivity, turnaround time, other co-alterations, and so on.12 For commercial kits, such as the FDA approved companion diagnostics (CDx), a verification in the local laboratory is needed. For the in-house methods, or so-called laboratory-developed tests (LDTs), an extensive validation must be performed to ensure the excellence of testing results. Because the percentage of tumor cells in the specimen directly affects the testing results, histological assessment of neoplastic cell content and surgical pathologist selection of the tissue block with the highest tumor content and best quality (to avoid the necrosis and hemorrhage areas), is the first crucial step for a successful testing.13 Macrodissection to enrich cancer cells in the low tumor content specimen is a practical method to increase the chance of testing success.
For patients without usable tumor tissue or for the purpose of follow up, liquid biopsy is a minimally invasive and rapid alternative that can capture comprehensive molecular changes. Liquid biopsy is also useful for early diagnosis, therapy response evaluation, and resistance mechanism exploration. Techniques like PCR-based methods and NGS are used to detect circulating tumor DNA (ctDNA) due to its low concentration and quality in plasma. The results of ctDNA testing may not always match the mutation profile of the primary tumors, possibly due to insufficient ctDNA release at early stages, differences in detection sensitivities, or tumor heterogeneity.14 Despite these challenges, liquid biopsy provides valuable complementary information to traditional tissue tests, though further large-scale studies are needed to standardize its clinical application.
It is critical for surgical and molecular pathologist to collaborate to select the most suitable platforms and tissue types to detect KRAS mutations. An accurate and timely result is the foundation of targeted KRAS inhibitor treatment.
KRAS inhibitors
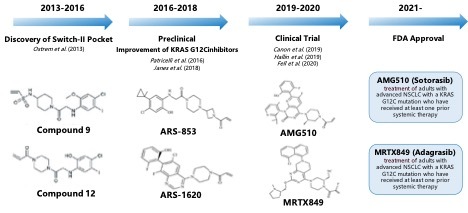
KRAS was considered undruggable due to its high resistance to small molecule modulation. The protein has a smooth surface with no well-defined pockets for inhibitor binding, except for its GTP/GDP-binding pocket, which is occupied by GTP with extremely high affinity. Given the high cellular concentration of GTP, designing a small molecule inhibitor to compete with GTP at this pocket was deemed formidable.15 In 2013, Ostrem, Shokat, and colleagues achieved a breakthrough by developing compounds that covalently and irreversibly bind to the cysteine residue of the KRAS G12C mutant.16 To date, two KRAS inhibitors targeting G12C mutants have received FDA approval: sotorasib and adagrasib. Additionally, several other drugs have advanced to clinical trials, including LY3499446, JAB-21822, JNJ-74699157, GFH925, YL-15293, MRTX-1133, and JDQ443.
Targeting KRAS (G12C)
The discovery of a new allosteric site in KRAS (G12C) has renewed interest in targeting this protein. The development of KRAS (G12C) inhibitors is illustrated in Figure 3. Researchers developed compounds like ARS-853 and ARS-1620 based on previous work. ARS-853 inhibits KRAS (G12C) by locking it in its inactive state, revealing that KRAS mutations vary biologically.17 ARS-1620, the first in vivo proof-of-concept molecule, binds more stably to KRAS (G12C) and is potent in various models. Despite weak binding affinity, the inhibitors are effective due to KRAS-mediated acceleration of the chemical reactions involved.18
AMG510 is the first small molecule inhibitor targeting KRAS (G12C) to enter clinical trials and has been approved by the FDA for treating NSCLC patients with this mutation. It binds specifically and irreversibly to Cys12, locking KRAS (G12C) in an inactive state, and shows tenfold improved efficiency over ARS-1620. Clinical trials showed promising results, with an overall response rate (ORR) of 37.1% and a disease control rate (DCR) of 80.6%, confirming its efficacy and safety.19 Ongoing trials are further assessing its potential in combination therapies and compared to other treatments. Adagrasib (MRTX849) is another FDA-approved selective inhibitor for KRAS-G12C-mutated NSCLC. It binds covalently to the GTP-bound KRAS G12C mutants’ SII-P region, inhibiting MAPK signaling and reducing cell viability at nanomolar concentrations. In vivo studies demonstrate strong dose-dependent inhibition of pERK and significant tumor regression, achieving complete regression in a pancreatic cancer xenograft model quicker than Sotorasib.20 The Krystal-1 Phase I trial showed a 43% objective response rate in NSCLC patients, with manageable adverse effects leading to discontinuation in only a few cases.21
Targeting other KRAS mutations
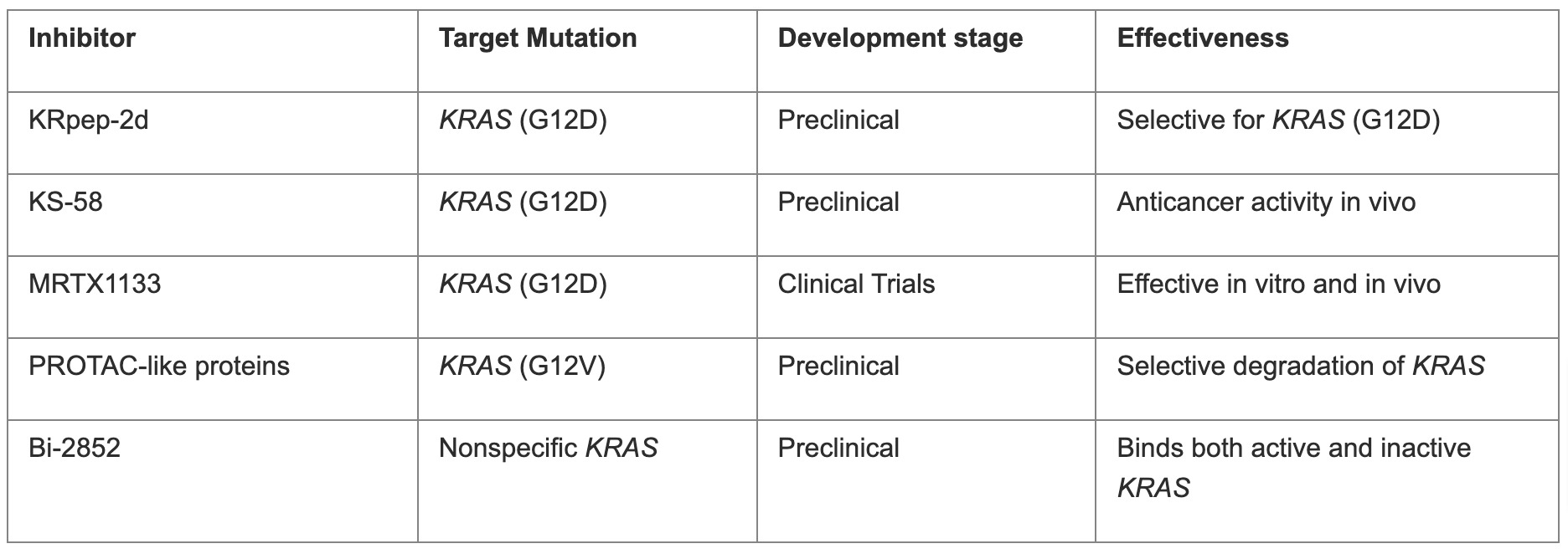
The development of KRAS (G12C) inhibitors relies on covalently inhibiting cysteine and high GTPase activity, traits not found in other KRAS mutations. To target other common KRAS mutations like KRAS (G12D) and KRAS (G12V), researchers are exploring new strategies. They identified a novel inhibitory peptide, KRpep-2d, selective for KRAS (G12D), and developed KS-58, which shows anticancer activity in vivo.22 MRTX1133 is a novel inhibitor that specifically targets the Asp12-mutated residue in G12D-mutated KRAS proteins, similar to the G12C inhibitor adagrasib. It effectively inhibited several G12D-mutated cell lines at nanomolar concentrations in vitro and demonstrated strong, dose-dependent tumor regression and repression of Erk signaling in vivo.23 While it showed less effectiveness against a single KRAS wild-type cell line in vitro, further research is needed to assess its selectivity against other KRAS mutants and wild-type backgrounds. MRTX1133 is the first inhibitor targeting KRAS (G12D) mutants entering clinical trials. PROTAC-like proteins were created to degrade KRAS (G12V) selectively. Kessler's team discovered Bi-2852, a nonspecific KRAS inhibitor that binds with high affinity to both active and inactive KRAS.23 Overall, while specific inhibitors for other KRAS mutations are still in early development, new approaches offer promise for future clinical applications. Table 4 provides a brief summary of current inhibitors targeting other KRAS mutations.
Table 4. Inhibitors targeting other KRAS mutations
Summary
Due to the high prevalence of KRAS mutations across cancer types, targeting KRAS is an appealing strategy. KRAS (G12C) inhibitors have been approved and utilized clinically. More drugs either targeting specific mutation or pan-KRAS inhibitor are under development or clinical trails. To meet the growing clinical need, rapid and accurate molecular detection is essential for guiding clinical decision-making. Pathologists are playing an increasingly important role in KRAS mutation detection to select the most suitable tissue block and sample type (as well as the best available and practical platform) to test KRAS gene mutations and provide guidance to treatment decisions, especially targeted KRAS inhibitor treatment.
References
- Jee J, Lebow ES, Yeh R, Das JP, et al. Overall survival with circulating tumor DNA-guided therapy in advanced non-small-cell lung cancer. Nat Med. 2002;28(11):2353-2363. doi: 10.1038/s41591-022-02047-z
- Li BT, Janku F, Jung B, et al. Ultra-deep next-generation sequencing of plasma cell-free DNA in patients with advanced lung cancers: results from the Actionable Genome Consortium. Ann Oncol. 2019;30(4):597-603. doi: 10.1093/annonc/mdz046
- Razavi P, Li BT, Brown DN, et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat Med. 2019;25(12):1928-1937. doi: 10.1038/s41591-019-0652-7
- Hunter JC, Manandhar A, Carrasco MA, et al. Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations. Mol Cancer Res. 2015;13(9):1325-1335. doi: 10.1158/1541-7786.MCR-15-0203
- Dogan S, Shen R, Ang DC, et al. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res. 2012;18(22):6169-77. doi: 10.1158/1078-0432.CCR-11-3265
- Haigis, KM. KRAS Alleles: The Devil Is in the Detail. Trends Cancer. 2017;3(10):686-697. doi: 10.1016/j.trecan.2017.08.006.
- Smith MJ, Neel BG, Ikura M. NMR-based functional profiling of RASopathies and oncogenic RAS mutations. Proc Natl Acad Sci U S A. 2013;110(12):4574-4579. doi: 10.1073/pnas.1218173110
- Ihle NT, Byers LA, Kim ES, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst. 2012;104(3):228-239. doi: 10.1093/jnci/djr523
- Linardou H, Briasoulis E, Dahabreh IJ, et al. All about KRAS for clinical oncology practice: gene profile, clinical implications and laboratory recommendations for somatic mutational testing in colorectal cancer. Cancer Treat Rev. 2011;37(3):221-233. doi: 10.1016/j.ctrv.2010.07.008
- Cross J. DxS Ltd. Pharmacogenomics. 2008;9:463-467.
- Gonzalez de Castro D, Angulo B, Gomez B, et al. A comparison of three methods for detecting KRAS mutations in formalin-fixed colorectal cancer specimens. Br J Cancer. 2012;107(2):345-351. doi: 10.1038/bjc.2012.259
- Herreros-Villanueva M, Chen CC, Yuan SS, et al. KRAS mutations: analytical considerations. Clin Chim Acta. 2014;431:211-220. doi: 10.1016/j.cca.2014.01.049
- Ross JS. Clinical implementation of KRAS testing in metastatic colorectal carcinoma: the pathologist's perspective. Arch Pathol Lab Med. 2012;136(10):1298-1307. doi: 10.5858/arpa.2011-0478-RA
- Zhu G, Pei L, Xia H, et al. Role of oncogenic KRAS in the prognosis, diagnosis and treatment of colorectal cancer. Mol Cancer. 2021;20(1):143. doi: 10.1186/s12943-021-01441-4
- Huang L, Guo Z, Wang F, Fu L. KRAS mutation: from undruggable to druggable in cancer. Signal Transduct Target Ther. 2021;6(1):386. doi: 10.1038/s41392-021-00780-4
- Ostrem JM, Shokat KM. Direct small-molecule inhibitors of KRAS: from structural insights to mechanism-based design. Nat Rev Drug Discov. 2016;15(11):771-785. doi: 10.1038/nrd.2016.139
- Patricelli MP, Janes MR, Li Ks, et al. Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discov. 2016;6(3):316-329. doi: 10.1158/2159-8290.CD-15-1105
- Janes MR, Zhang J, Li LS, et al. Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell. 2018;172(3):578-589. doi: 10.1016/j.cell.2018.01.006
- Hong DS, Fakih MG, Strickler JH, et al. KRAS(G12C) Inhibition with sotorasib in advanced solidtTumors. N Engl J Med. 2020;383(13):1207-1217. doi: 10.1056/NEJMoa1917239
- Fell JB, Fischer JP, Baer BR, et al. Identification of the clinical development candidate MRTX849, a covalent KRAS(G12C) inhibitor for the treatment of vancer. J Med Chem. 2020;63(13):6679-6693. doi: 10.1021/acs.jmedchem.9b02052
- Jänne PA, Riely GJ, Gadgeel SM, et al. Adagrasib in non-small-cell lung cancer harboring a KRAS(G12C) mutation. N Engl J Med. 2022;387(2):120-131. doi: 10.1056/NEJMoa2204619
- Sakamoto K, Kamada Y, Sameshima T, et al. K-Ras(G12D)-selective inhibitory peptides generated by random peptide T7 phage display technology. Biochem Biophys Res Commun. 2017;484(3): 605-611. doi: 10.1016/j.bbrc.2017.01.147
- Wang X, Allen S, Blake JF, et al. Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS(G12D) Inhibitor. J Med Chem. 2022;65(4):3123-3133. doi: 10.1021/acs.jmedchem.1c01688

Jinjuan Yao, MD, PhD, FCAP, is a molecular pathologist from Memorial Sloan Kettering Cancer Center (MSK) with subspecialty training in molecular pathology and hematopathology. Dr. Yao also serves as the Quality Assurance Chair of Diagnostic Molecular Pathology at MSK.
Dr. Yao is a lifetime member of Chinese American Pathologists Association (CAPA) and the chair for CAPA molecular pathology subcommittee. She is also a CAP New York House of Delegates and CAP Personalized Health Care Committee (PHC) member.
Dr. Yao’s clinical work has focused on the large-scale, prospective genotyping of solid tumors and hematologic malignancies using a variety of NGS technologies. Her research interests include the pathogenesis of leukemias and lymphomas, genetic alterations, and therapies of solid tumors.

Jing Wang, MD, is an attending physician in Department of Pathology, Peking Union Medical College Hospital. She is currently a visiting investigator in Memorial Sloan Kettering Cancer Center. She received her MD degree at Peking Union Medical College. Her areas of interests include use of next-generation sequencing technologies for tumor diagnosis and biomarker discovery. Her clinical research has been focusing on tumor heterogeneity and tumor immune microenvironment of colorectal cancer, endometrial cancer and breast cancer.